7Q0O
 
 | | E. coli NfsA | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NADPH nitroreductase | | Authors: | White, S.A, Grainger, A, Parr, R, Day, M.A, Jarrom, D, Graziano, A, Searle, P.F, Hyde, E.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | The 3D-structure, kinetics and dynamics of the E. coli nitroreductase NfsA with NADP + provide glimpses of its catalytic mechanism.
Febs Lett., 596, 2022
|
|
4U6N
 
 | | Crystal structure of Escherichia coli DiaA | | Descriptor: | CHLORIDE ION, DnaA initiator-associating protein DiaA | | Authors: | Oakley, A.J, Lo, T. | | Deposit date: | 2014-07-29 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of Escherichia coli DiaA
To Be Published
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
4UDS
 
 | | Crystal structure of MbdR regulator from Azoarcus sp. CIB | | Descriptor: | MBDR REGULATOR | | Authors: | Liu, H, Liu, H.X, MacMahon, S.A, Juarez, J.F, Zamarro, M.T, Eberlein, C, Boll, M, Carmona, M, Diaz, E, Naismith, J.H. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unraveling the Specific Regulation of the Mbd Central Pathway for the Anaerobic Degradation of 3-Methylbenzoate.
J.Biol.Chem., 290, 2015
|
|
7PDY
 
 | | A viral peptide from Marek's disease virus bound to chicken MHC-II molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 38 kDa phosphoprotein,MHC class II beta chain, ... | | Authors: | Goryanin, A, Cook, A.G, Kaufman, J, Halabi, S. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Viral peptide bound to chicken MHC-II molecule
To Be Published
|
|
7PDV
 
 | |
6P2K
 
 | | Crystal structure of AFV00434, an ancestral GH74 enzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
6MTP
 
 | | Crystal structure of VRC42.04 Fab in complex with gp41 peptide | | Descriptor: | Antibody VRC42.04 Fab heavy chain, Antibody VRC42.04 Fab light chain, RV217 founder virus gp41 peptide | | Authors: | Kwon, Y.D, Druz, A, Law, W.H, Peng, D, Zhang, B, Doria-Rose, N.A, Kwong, P.D. | | Deposit date: | 2018-10-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.036 Å) | | Cite: | Longitudinal Analysis Reveals Early Development of Three MPER-Directed Neutralizing Antibody Lineages from an HIV-1-Infected Individual.
Immunity, 50, 2019
|
|
7NBP
 
 | | Solution structure of DNA duplex containing a 7,8-dihydro-8-oxo-1,N6-ethenoadenine base modification that induces exclusively A->T transversions in Escherichia coli | | Descriptor: | DNA (5'-D(*CP*AP*TP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*(EDA)P*CP*AP*TP*G)-3') | | Authors: | Aralov, A.V, Gubina, N, Cabrero, C, Tsvetkov, V.B, Turaev, A.V, Fedeles, B.I, Croy, R.G, Isaakova, E.A, Melnik, D, Dukova, S, Khrulev, A.A, Varizhuk, A.M, Gonzalez, C, Zatsepin, T.S, Essigmann, J.M. | | Deposit date: | 2021-01-27 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 7,8-Dihydro-8-oxo-1,N6-ethenoadenine: an exclusively Hoogsteen-paired thymine mimic in DNA that induces A→T transversions in Escherichia coli.
Nucleic Acids Res., 50, 2022
|
|
7PNL
 
 | | Complex between monomolecular human telomeric G-quadruplex and a sulfonamide derivative of the natural alkaloid Berberine | | Descriptor: | 4-[[1-[3-[(17-methoxy-5,7-dioxa-13-azoniapentacyclo[11.8.0.0^{2,10}.0^{4,8}.0^{15,20}]henicosa-1(21),2(10),3,8,13,15,17,19-octaen-16-yl)oxy]propyl]triazol-4-yl]methoxy]benzenesulfonamide, G-guadruplex DNA (23-mer), POTASSIUM ION | | Authors: | Bazzicalupi, C, Gratteri, P, Petreni, A, Nocentini, A. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Development of a multi-targeted chemotherapeutic approach based on G-quadruplex stabilisation and carbonic anhydrase inhibition.
J Enzyme Inhib Med Chem, 39, 2024
|
|
8EXA
 
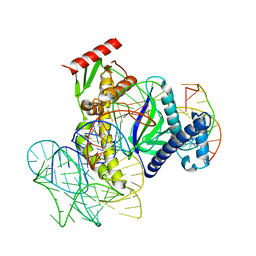 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8EX9
 
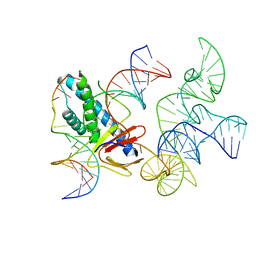 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
4UID
 
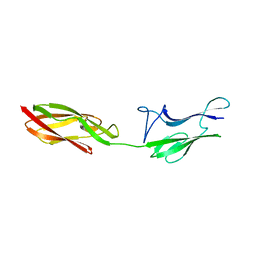 | |
6P2O
 
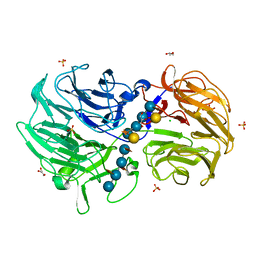 | | Crystal structure of Streptomyces rapamycinicus GH74 in complex with xyloglucan fragments XLLG and XXXG | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity, regiospecificity, and processivity in glycoside hydrolase family 74.
J.Biol.Chem., 294, 2019
|
|
8EYW
 
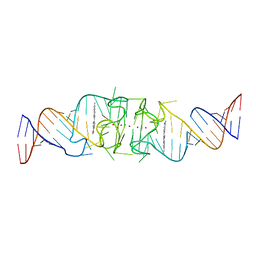 | | Beetroot dimer bound to ThT | | Descriptor: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
7Q1D
 
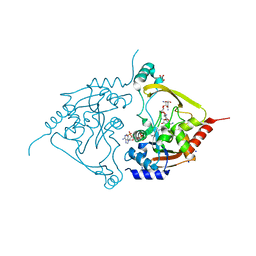 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
8EYU
 
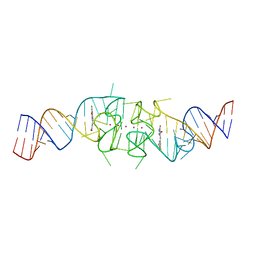 | | Structure of Beetroot dimer bound to DFAME | | Descriptor: | POTASSIUM ION, RNA (49-MER), methyl (2E)-3-{(4Z)-4-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-1-methyl-5-oxo-4,5-dihydro-1H-imidazol-2-yl}prop-2-enoate | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
7Q46
 
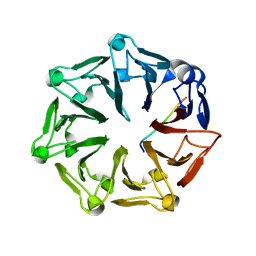 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Pericentriolar material 1 protein | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.46002531 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of pericentriolar material 1 protein
To Be Published
|
|
7Q43
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10 | | Descriptor: | CITRIC ACID, Dedicator of cytokinesis protein 10 peptide, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40002346 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of dedicator of cytokinesis protein 10
To Be Published
|
|
8EYV
 
 | | Structure of Beetroot dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (45-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-10-28 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
8F0N
 
 | | Wobble Beetroot (A16U-U38G) dimer bound to DFHO | | Descriptor: | (5Z)-5-[(3,5-difluoro-4-hydroxyphenyl)methylidene]-2-[(E)-(hydroxyimino)methyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, POTASSIUM ION, RNA (49-MER) | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-11-03 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Co-crystal structures of the fluorogenic aptamer Beetroot show that close homology may not predict similar RNA architecture.
Nat Commun, 14, 2023
|
|
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
7Q41
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP) | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Ubiquitin-protein ligase E3A (E6AP) peptide | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.01478052 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP)
To Be Published
|
|
7Q45
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Myelin transcription factor 1 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09999585 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1
To Be Published
|
|
7Q42
 
 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95002484 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B
To Be Published
|
|
