3LS0
 
 | |
4ZY8
 
 | |
7BIK
 
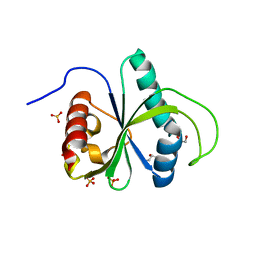 | | Crystal structure of YTHDF2 in complex with m6Am | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X, Caflisch, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
5FP2
 
 | |
1N4X
 
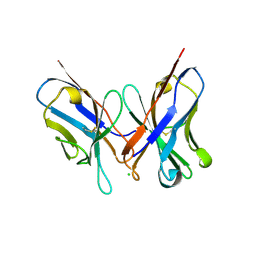 | | Structure of scFv 1696 at acidic pH | | Descriptor: | CHLORIDE ION, immunoglobulin heavy chain variable region, immunoglobulin kappa chain variable region | | Authors: | Lescar, J, Brynda, J, Fabry, M, Horejsi, M, Rezacova, P, Sedlacek, J, Bentley, G.A. | | Deposit date: | 2002-11-02 | | Release date: | 2003-06-10 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a single-chain Fv fragment of an antibody that inhibits the HIV-1 and HIV-2 proteases.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5FH2
 
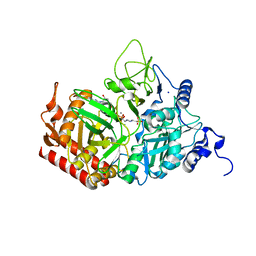 | | The structure of rat cytosolic PEPCK variant E89Q in complex with GTP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Johnson, T.A, Holyoak, T. | | Deposit date: | 2015-12-21 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Utilization of Substrate Intrinsic Binding Energy for Conformational Change and Catalytic Function in Phosphoenolpyruvate Carboxykinase.
Biochemistry, 55, 2016
|
|
4EJE
 
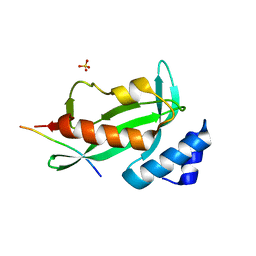 | |
4ZVU
 
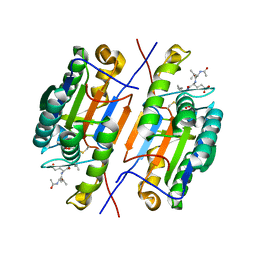 | |
5FMS
 
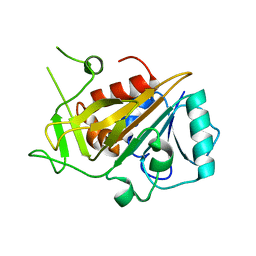 | | mmIFT52 N-terminal domain | | Descriptor: | INTRAFLAGELLAR TRANSPORT PROTEIN 52 HOMOLOG | | Authors: | Mourao, A, Lorentzen, E. | | Deposit date: | 2015-11-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.488 Å) | | Cite: | Intraflagellar Transport Proteins 172, 80, 57, 54, 38, and 20 Form a Stable Tubulin-Binding Ift-B2 Complex.
Embo J., 35, 2016
|
|
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6ECK
 
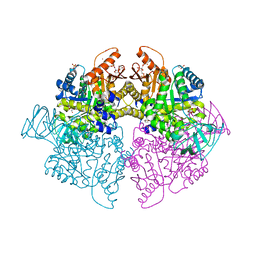 | | Pyruvate Kinase Isoform L-type with phosphorylated Ser113 (pS113) in complex with FBP | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, CITRATE ANION, ... | | Authors: | Padyana, A, Tong, S. | | Deposit date: | 2018-08-08 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Distinct Hepatic PKA and CDK Signaling Pathways Control Activity-Independent Pyruvate Kinase Phosphorylation and Hepatic Glucose Production.
Cell Rep, 29, 2019
|
|
7KCV
 
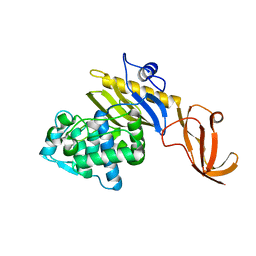 | |
7KCY
 
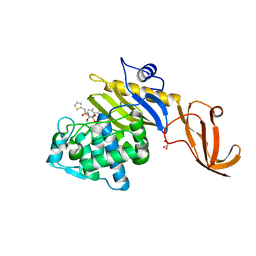 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | Descriptor: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
3OOU
 
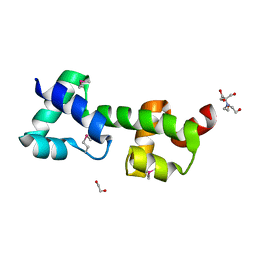 | | The structure of a protein with unkown function from Listeria innocua | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lin2118 protein | | Authors: | Fan, Y, Mack, J, Feldman, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The structure of a protein with unkown function from Listeria innocua
To be Published
|
|
5VJV
 
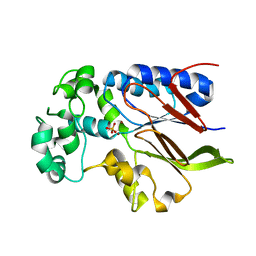 | | Rhizobiales-like phosphatase 2 | | Descriptor: | PHOSPHATE ION, Rhizobiales-like phosphatase 2, ZINC ION | | Authors: | Ng, K.K.S, Labandera, A, Moorhead, G. | | Deposit date: | 2017-04-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the preference of the Arabidopsis thalianaphosphatase RLPH2 for tyrosine-phosphorylated substrates.
Sci Signal, 11, 2018
|
|
3OP9
 
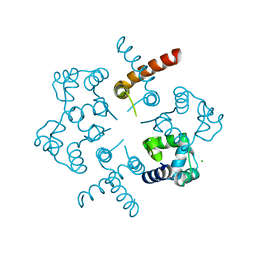 | |
7BFA
 
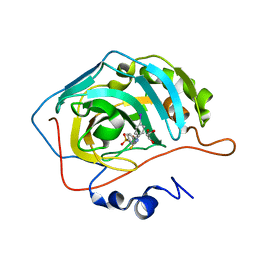 | |
7YKE
 
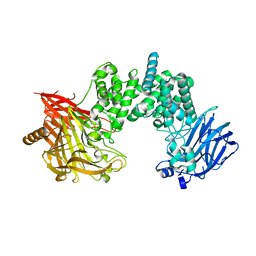 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 4,6-sulfate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Watanabe, I, Miyanaga, A, Eguchi, T. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Biochemical and crystallographic assessments of the effect of 4,6-O-disulfated disaccharide moieties in chondroitin sulfate E on chondroitinase ABC I activity.
Febs J., 290, 2023
|
|
5A7H
 
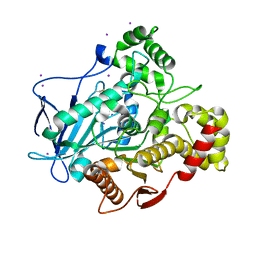 | | Comparison of the structure and activity of glycosylated and aglycosylated Human Carboxylesterase 1 | | Descriptor: | IODIDE ION, LIVER CARBOXYLESTERASE 1 | | Authors: | Arena de Souza, V, Scott, D.J, Charlton, M, Walsh, M.A, Owen, R.J. | | Deposit date: | 2015-07-04 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparison of the Structure and Activity of Glycosylated and Aglycosylated Human Carboxylesterase 1.
Plos One, 10, 2015
|
|
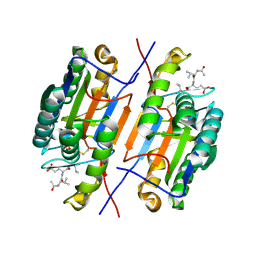
4ZVQ
 
 | |
3OPU
 
 | |
7NEX
 
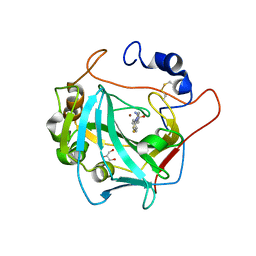 | | Crystal structure of alpha Carbonic anhydrase from Schistosoma mansoni bound to 1-(4-fluorophenyl)-3-(4-sulfamoylph enyl)thiourea | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2021-02-05 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
4MR4
 
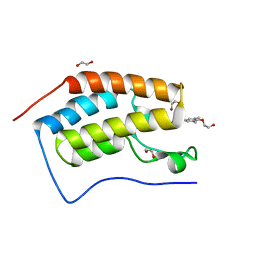 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolinone ligand (RVX-208) | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(2-hydroxyethoxy)-3,5-dimethylphenyl]-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3BYW
 
 | | Crystal structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae | | Descriptor: | ACETATE ION, Putative arabinofuranosyltransferase, ZINC ION | | Authors: | Tan, K, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of an extracellular domain of arabinofuranosyltransferase from Corynebacterium diphtheriae.
To be Published
|
|
7NG1
 
 | | Crystal structure of alpha Carbonic anhydrase from Schistoso ma mansoni bound to 1-(4-iodophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]selenourea | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carbonic anhydrase, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2021-02-08 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Insights into Schistosoma mansoni Carbonic Anhydrase (SmCA) Inhibition by Selenoureido-Substituted Benzenesulfonamides.
J.Med.Chem., 64, 2021
|
|
