5KVG
 
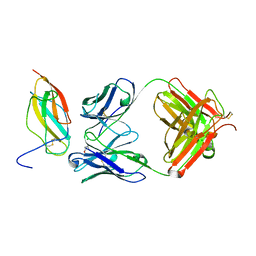 | | Zika specific antibody, ZV-67, bound to ZIKA envelope DIII | | Descriptor: | CHLORIDE ION, ZIKA Envelope DIII, ZV-67 Antibody Fab Heavy Chain, ... | | Authors: | Zhao, H, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-07-14 | | Release date: | 2016-08-03 | | Last modified: | 2016-08-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Zika Virus-Specific Antibody Protection.
Cell, 166, 2016
|
|
7LZX
 
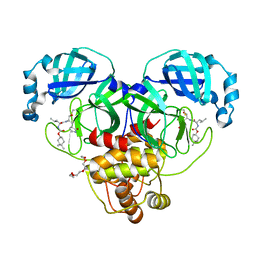 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 1c | | Descriptor: | (1R,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((4,4-dimethylcyclohexyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
4QGZ
 
 | |
7LZU
 
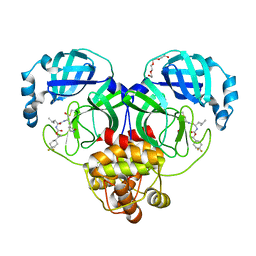 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 12b | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)ethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
7ZGT
 
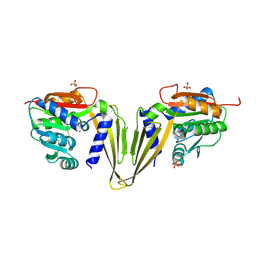 | | C-Methyltransferase PsmD from Streptomyces griseofuscus (apo form) | | Descriptor: | FORMIC ACID, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Weiergraeber, O.H, Amariei, D.A, Pozhydaieva, N, Pietruszka, J. | | Deposit date: | 2022-04-04 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic C3-Methylation of Indoles Using Methyltransferase PsmD-Crystal Structure, Catalytic Mechanism, and Preparative Applications
Acs Catalysis, 2022
|
|
5V7V
 
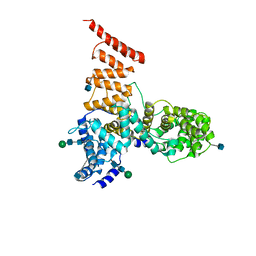 | | Cryo-EM structure of ERAD-associated E3 ubiquitin-protein ligase component HRD3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ERAD-associated E3 ubiquitin-protein ligase component HRD3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Mi, W, Schoebel, S, Stein, A, Rapoport, T.A, Liao, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-08-16 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3.
Nature, 548, 2017
|
|
7M04
 
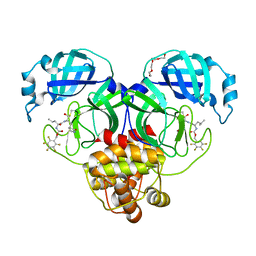 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 21c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-((((perfluorophenyl)methoxy)carbonyl)amino)pentanamido)-3-((R)-2-oxo-2,3-dihydro-1H-pyrrol-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
5V8D
 
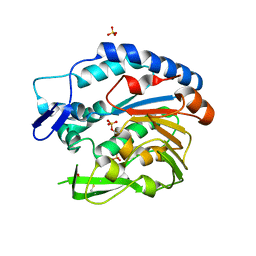 | | Structure of Bacillus cereus PatB1 with sulfonyl adduct | | Descriptor: | Bacillus cereus PatB1, SULFATE ION | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
4QLV
 
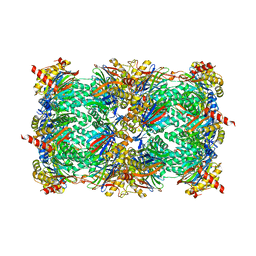 | | yCP in complex with tripeptidic epoxyketone inhibitor 17 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(morpholin-4-ylacetyl)-D-alanyl-N-[(2S,4R)-5-hydroxy-4-methyl-3-oxo-1-phenylpentan-2-yl]-O-methyl-L-tyrosinamide, ... | | Authors: | de Bruin, G, Huber, E, Xin, B, van Rooden, E, Al-Ayed, K, Kim, K, Kisselev, A, Driessen, C, van der Marel, G, Groll, M, Overkleeft, H. | | Deposit date: | 2014-06-13 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-based design of beta 1i or beta 5i specific inhibitors of human immunoproteasomes
J.Med.Chem., 57, 2014
|
|
4YXR
 
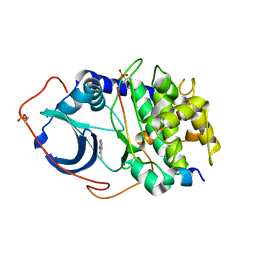 | | CRYSTAL STRUCTURE OF PKA IN COMPLEX WITH inhibitor. | | Descriptor: | 3-methyl-2H-indazole, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Schiffer, A, Wendt, K.U. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A combination of spin diffusion methods for the determination of protein-ligand complex structural ensembles.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7SQI
 
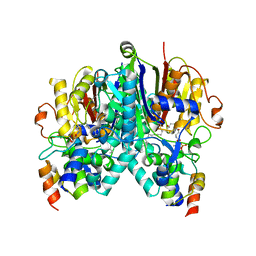 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | Descriptor: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | Authors: | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-05 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5N24
 
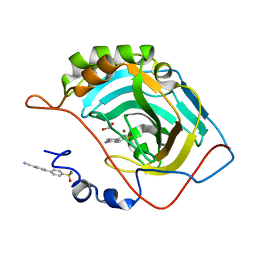 | | Crystal structure of human carbonic anhydrase II in complex with the inhibitor b4'-Cyano-biphenyl-4-sulfonic acid amide | | Descriptor: | 4-(4-cyanophenyl)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Supuran, C.T, Scozzafava, A, Carta, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfonamide carbonic anhydrase inhibitors: Zinc coordination and tail effects influence inhibitory efficacy and selectivity for different isoforms
Inorg.Chim.Acta., 470, 2018
|
|
4YZ3
 
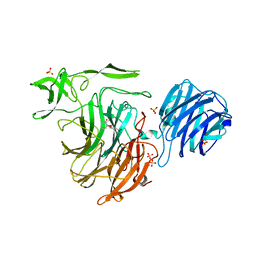 | | Crystal Structure of Streptococcus pneumoniae NanC, in complex with Oseltamivir. | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase, SULFATE ION | | Authors: | Lukacik, P, Owen, C.D, Potter, J.A, Taylor, G.L, Walsh, M.A. | | Deposit date: | 2015-03-24 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal Structure of Streptococcus pneumoniae NanC.
To Be Published
|
|
6T1J
 
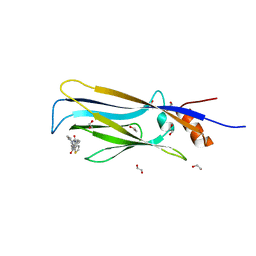 | | Crystal structure of MLLT1 (ENL) YEATS domain in complexed with piperazine-urea derivative 2 | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL, ~{N}-[[4-(pyrrolidin-1-ylmethyl)phenyl]methyl]-4-thiophen-2-ylcarbonyl-piperazine-1-carboxamide | | Authors: | Chaikuad, A, Heidenreich, D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Insights into Interaction Mechanisms of Alternative Piperazine-urea YEATS Domain Binders in MLLT1.
Acs Med.Chem.Lett., 10, 2019
|
|
8SWS
 
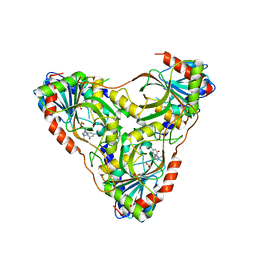 | | Structure of K. lactis PNP S42E-H98R variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
5V9J
 
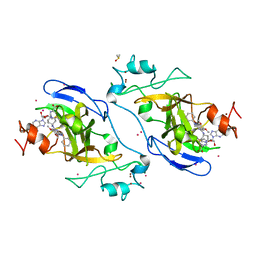 | | Crystal structure of catalytic domain of GLP with MS0105 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase EHMT1, ... | | Authors: | Dong, A, Zeng, H, Liu, J, Xiong, Y, Babault, N, Jin, J, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Wu, H, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-23 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of catalytic domain of GLP with MS0105
to be published
|
|
7C3N
 
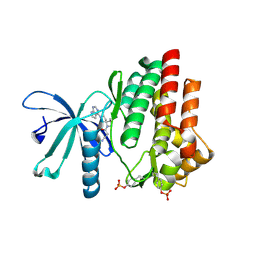 | | Crystal structure of JAK3 in complex with Delgocitinib | | Descriptor: | 3-[(3S,4R)-3-methyl-7-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)-1,7-diazaspiro[3.4]octan-1-yl]-3-oxidanylidene-propanenitrile, Tyrosine-protein kinase JAK3 | | Authors: | Doi, S, Otira, T, Kikuwaka, M, Nomura, A, Noji, S, Adachi, T. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Janus Kinase Inhibitor Bearing a Highly Three-Dimensional Spiro Scaffold: JTE-052 (Delgocitinib) as a New Dermatological Agent to Treat Inflammatory Skin Disorders.
J.Med.Chem., 63, 2020
|
|
4Z0X
 
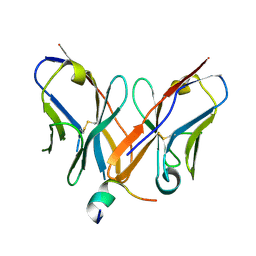 | |
5VBG
 
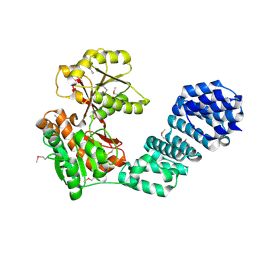 | |
6DIZ
 
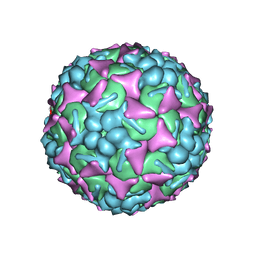 | | EV-A71 strain 11316 complexed with tryptophan dendrimer MADAL_0385 | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Sun, L, Lee, H, Thibaut, H.J, Rivero-Buceta, E, Martinez-Gualda, B, Delang, L, Leyssen, P, Gago, F, San-Felix, A, Hafenstein, S, Mirabelli, C, Neyts, J. | | Deposit date: | 2018-05-24 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Viral engagement with host (co-)receptors blocked by a novel class of tryptophan dendrimers that targets the 5-fold-axis of the enterovirus-A71 capsid.
Plos Pathog., 15, 2019
|
|
6FSP
 
 | | Crystal structure of APRT from Thermus thermophilus | | Descriptor: | PRPP-binding protein, adenine/guanine phosphoribosyltransferase | | Authors: | Timofeev, V.I, Sinitsyna, E.V, Kostromina, M.A, Muravieva, T.I, Makarov, D.A, Mikheeva, O.O, Kuranova, I.P, Esipov, R.S. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of APRT from Thermus thermophilus
To Be Published
|
|
4QMH
 
 | | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array | | Descriptor: | LP04448p, SULFATE ION | | Authors: | Fox, J.C, Howard, A.E, Currie, J.D, Rogers, S.L, Slep, K.C. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array.
Mol.Biol.Cell, 25, 2014
|
|
5DZH
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | Descriptor: | 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
4Z0K
 
 | |
4QG4
 
 | | Crystal structure of the tetrameric GTP/dATP/ATP-bound SAMHD1 (H210A) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
