1TLM
 
 | |
2IFU
 
 | | Crystal Structure of a Gamma-SNAP from Danio rerio | | Descriptor: | SULFATE ION, gamma-snap | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-09-21 | | Release date: | 2006-10-10 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and dynamics of gamma-SNAP: insight into flexibility of proteins from the SNAP family.
Proteins, 70, 2008
|
|
1T13
 
 | | Crystal Structure Of Lumazine Synthase From Brucella Abortus Bound To 5-nitro-6-(D-ribitylamino)-2,4(1H,3H) pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Vega, D.R, Guimaraes, B.G, Braden, B.C, Goldbaum, F.A. | | Deposit date: | 2004-04-15 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies on Decameric Brucella spp. Lumazine Synthase: A Novel Quaternary Arrangement Evolved for a New Function?
J.Mol.Biol., 353, 2005
|
|
2IK6
 
 | | Yeast inorganic pyrophosphatase variant D120E with magnesium and phosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Inorganic pyrophosphatase, MAGNESIUM ION, ... | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IK9
 
 | | Yeast inorganic pyrophosphatase variant D152E with magnesium and phosphate | | Descriptor: | Inorganic pyrophosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Oksanen, E, Ahonen, A.K, Tuominen, H, Tuominen, V, Lahti, R, Goldman, A, Heikinheimo, P. | | Deposit date: | 2006-10-02 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Complete Structural Description of the Catalytic Cycle of Yeast Pyrophosphatase.
Biochemistry, 46, 2007
|
|
2IL4
 
 | | Crystal structure of At1g77540-Coenzyme A Complex | | Descriptor: | COENZYME A, Protein At1g77540 | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.054 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1TKD
 
 | | T7 DNA polymerase ternary complex with 8 oxo guanosine and dCMP at the elongation site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*CP*CP*CP*AP*(8OG)P*TP*GP*GP*CP*AP*CP*TP*GP*GP*CP*CP*GP*TP*CP*GP*TP*TP*TP*TP*CP*G)-3'), ... | | Authors: | Brieba, L.G, Eichman, B.F, Kokoska, R.J, Doublie, S, Kunkel, T.A, Ellenberger, T. | | Deposit date: | 2004-06-08 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase.
Embo J., 23, 2004
|
|
3F0A
 
 | | Structure of a putative n-acetyltransferase (ta0374) in complex with acetyl-coa from thermoplasma acidophilum | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, N-ACETYLTRANSFERASE, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3F1Q
 
 | | Human dihydroorotate dehydrogenase in complex with a leflunomide derivative inhibitor 1 | | Descriptor: | (2Z)-N-biphenyl-4-yl-2-cyano-3-hydroxybut-2-enamide, ACETIC ACID, Dihydroorotate dehydrogenase, ... | | Authors: | Heikkila, T, Davies, M, McConkey, G.A, Fishwick, C.W.G, Parsons, M.R, Johnson, A.P. | | Deposit date: | 2008-10-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and characterization of inhibitors of human and Plasmodium falciparum dihydroorotate dehydrogenases
J.Med.Chem., 52, 2009
|
|
1T2E
 
 | | Plasmodium falciparum lactate dehydrogenase S245A, A327P mutant complexed with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, L-lactate dehydrogenase, ... | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-21 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
1T26
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with NADH and 4-hydroxy-1,2,5-thiadiazole-3-carboxylic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-HYDROXY-1,2,5-THIADIAZOLE-3-CARBOXYLIC ACID, L-lactate dehydrogenase | | Authors: | Cameron, A, Read, J, Tranter, R, Winter, V.J, Sessions, R.B, Brady, R.L, Vivas, L, Easton, A, Kendrick, H, Croft, S.L, Barros, D, Lavandera, J.L, Martin, J.J, Risco, F, Garcia-Ochoa, S, Gamo, F.J, Sanz, L, Leon, L, Ruiz, J.R, Gabarro, R, Mallo, A, De Las Heras, F.G. | | Deposit date: | 2004-04-20 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and Activity of a Series of Azole-based Compounds with Lactate Dehydrogenase-directed Anti-malarial Activity.
J.Biol.Chem., 279, 2004
|
|
8QIG
 
 | | CrPhotLOV1 light state structure 17.5 ms (15-20 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography
Iucrj, 11, 2024
|
|
2I06
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With DNA- Locked form | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*G*AP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli.
To be Published
|
|
8QIO
 
 | | CrPhotLOV1 light state structure 52.5 ms (50-55 ms) after illumination determined by time-resolved serial synchrotron crystallography at room temperature | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin | | Authors: | Gotthard, G, Mous, S, Weinert, T, Maia, R.N.A, James, D, Dworkowski, F, Gashi, D, Antonia, F, Wang, M, Panepucci, E, Ozerov, D, Schertler, G.F.X, Heberle, J, Standfuss, J, Nogly, P. | | Deposit date: | 2023-09-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Capturing the blue-light activated state of the Phot-LOV1 domain from Chlamydomonas reinhardtii using time-resolved serial synchrotron crystallography
Iucrj, 11, 2024
|
|
7S83
 
 | | Crystal structure of SARS CoV-2 Spike Receptor Binding Domain in complex with shark neutralizing VNARs ShAb01 and ShAb02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ShAb01 VNAR, ... | | Authors: | Chen, W.-H, Hajduczki, A, Dooley, H.M, Joyce, M.G. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Shark nanobodies with potent SARS-CoV-2 neutralizing activity and broad sarbecovirus reactivity.
Nat Commun, 14, 2023
|
|
4R2X
 
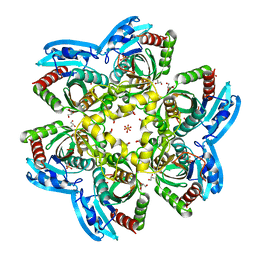 | | Unique conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis uridine phosphorylase in the free form and in complex with uridine | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Safonova, T.N, Mordkovich, N.N, Manuvera, V.A, Veiko, V.P, Popov, V.O, Polyakov, K.M. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | High-syn conformation of uridine and asymmetry of the hexameric molecule revealed in the high-resolution structures of Shewanella oneidensis MR-1 uridine phosphorylase in the free form and in complex with uridine.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2I4I
 
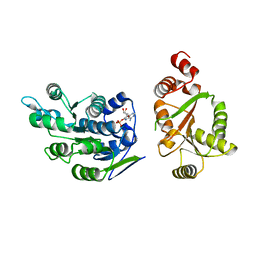 | | Crystal Structure of human DEAD-box RNA helicase DDX3X | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent RNA helicase DDX3X | | Authors: | Hogbom, M, Karlberg, T, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Conserved Domains 1 and 2 of the Human DEAD-box Helicase DDX3X in Complex with the Mononucleotide AMP
J.Mol.Biol., 372, 2007
|
|
1JJX
 
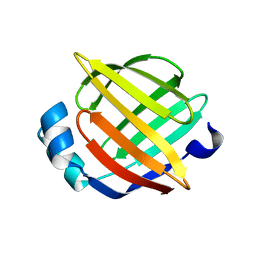 | | Solution Structure of Recombinant Human Brain-type Fatty acid Binding Protein | | Descriptor: | BRAIN-TYPE FATTY ACID BINDING PROTEIN | | Authors: | Rademacher, M, Zimmerman, A.W, Rueterjans, H, Veerkamp, J.H, Luecke, C. | | Deposit date: | 2001-07-10 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of fatty acid-binding protein from human brain.
Mol.Cell.Biochem., 239, 2002
|
|
3EOT
 
 | |
1TBQ
 
 | |
2HYU
 
 | | Human Annexin A2 with heparin tetrasaccharide bound | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Annexin A2, CALCIUM ION | | Authors: | Shao, C, Head, J.F, Seaton, B.A. | | Deposit date: | 2006-08-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystallographic Analysis of Calcium-dependent Heparin Binding to Annexin A2.
J.Biol.Chem., 281, 2006
|
|
1SXU
 
 | | 1.4 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Imidazole | | Descriptor: | IMIDAZOLE, Nitrophorin 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
6PDQ
 
 | | Ancestral Effector Caspase 3/6/7 | | Descriptor: | Ac-DEVD inhibitor, Ancestral Effector Caspase-3/6/7 | | Authors: | Clark, A.C. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Resurrection of ancestral effector caspases identifies novel networks for evolution of substrate specificity.
Biochem.J., 476, 2019
|
|
7ZV5
 
 | |
1SY3
 
 | | 1.00 A Crystal Structure of D30N Mutant of Nitrophorin 4 from Rhodnius Prolixus Complexed with Nitric Oxide | | Descriptor: | NITRIC OXIDE, Nitrophorin 4, PHOSPHATE ION, ... | | Authors: | Maes, E.M, Weichsel, A, Andersen, J.F, Shepley, D, Montfort, W.R. | | Deposit date: | 2004-03-31 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4
Biochemistry, 43, 2004
|
|
