5BP4
 
 | | Modifying region (DH-ER-KR) of a mycocerosic acid synthase-like (MAS-like) PKS | | Descriptor: | Mycocerosic acid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Herbst, D.A, Jakob, P.R, Zaehringer, F, Maier, T. | | Deposit date: | 2015-05-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases.
Nature, 531, 2016
|
|
6R5X
 
 | |
8DJ0
 
 | | Crystal structure of NavAb L123T as a basis for the human Nav1.7 Inherited Erythromelalgia I848T mutation | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for severe pain caused by mutations in the S4-S5 linkers of voltage-gated sodium channel Na V 1.7.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6R64
 
 | | N-terminal domain of modification dependent EcoKMcrA restriction endonuclease (NEco) in complex with C5mCGG target sequence | | Descriptor: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*CP*(5CM)P*GP*GP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*CP*(5CM)P*GP*GP*TP*TP*C)-3') | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
6R67
 
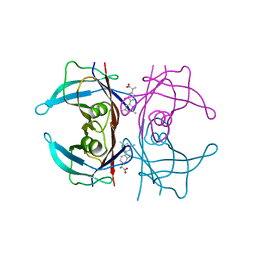 | | Crystal structure of transthyretin in complex with CHF5075, a flurbiprofen analogue | | Descriptor: | (2~{R})-2-[4-(3,5-dimethylphenyl)-3-fluoranyl-phenyl]propanoic acid, Transthyretin | | Authors: | Loconte, V, Menozzi, I, Ferrari, A, Berni, R, Zanotti, G. | | Deposit date: | 2019-03-26 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-activity relationships of flurbiprofen analogues as stabilizers of the amyloidogenic protein transthyretin.
J.Struct.Biol., 208, 2019
|
|
7QCP
 
 | |
6DWF
 
 | |
5FNG
 
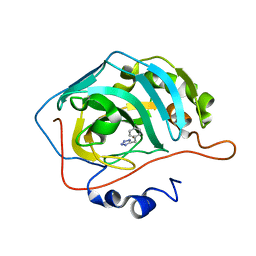 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(4-chlorophenyl)methyl]-1,2,3-triaza-4-azanidacyclopenta-2,5-diene, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-11-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
7JIU
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
5FV9
 
 | | Crystal structure of GalNAc-T2 in complex with compound 16d | | Descriptor: | 1,2-ETHANEDIOL, GALNAC-T2, GLYCEROL, ... | | Authors: | Ghirardello, M, Rivas, M, Lacetera, A, Delso, I, Lira-Navarrete, E, Tejero, T, Martin-Santamaria, S, Hurtado-Guerrero, R, Merino, P. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Glycomimetics Targeting Glycosyltransferases: Synthetic, Computational and Structural Studies of Less-Polar Conjugates.
Chemistry, 22, 2016
|
|
8JHE
 
 | | Hyper-thermostable ancestral L-amino acid oxidase 2 (HTAncLAAO2) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hyper thermostable ancestral L-amino acid oxidase | | Authors: | Kawamura, Y, Ishida, C, Miyata, R, Miyata, A, Hayashi, S, Fujinami, D, Ito, S, Nakano, S. | | Deposit date: | 2023-05-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of hyper-thermostable ancestral L-amino acid oxidase that can convert Trp derivatives to D-forms by chemoenzymatic reaction.
Commun Chem, 6, 2023
|
|
6RIE
 
 | | Structure of Vaccinia Virus DNA-dependent RNA polymerase co-transcriptional capping complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Hillen, H.S, Bartuli, J, Grimm, C, Dienemann, C, Bedenk, K, Szalar, A, Fischer, U, Cramer, P. | | Deposit date: | 2019-04-23 | | Release date: | 2019-12-18 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
5W9R
 
 | | Toxoplasma Gondii CDPK1 in complex with inhibitor LJQ138 | | Descriptor: | 1-tert-butyl-3-[(1H-indol-3-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Lovato, D, Loppnau, P, Lin, Y.H, Rutaganaria, F, Lopez, M.S, Shokat, L, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Sibley, D, Hui, R, Walker, J.R. | | Deposit date: | 2017-06-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Toxoplasma Gondii CDPK1 in complex with inhibitor LJQ138
To be published
|
|
7ZOM
 
 | |
5IWP
 
 | | Structure of Transient Receptor Potential (TRP) channel TRPV6 in the presence of calcium | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Saotome, K, Singh, A.K, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2016-03-22 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Crystal structure of the epithelial calcium channel TRPV6.
Nature, 534, 2016
|
|
5FT1
 
 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ bound to RNase E of Pseudomonas aeruginosa | | Descriptor: | GP37, RIBONUCLEASE E | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
8DHE
 
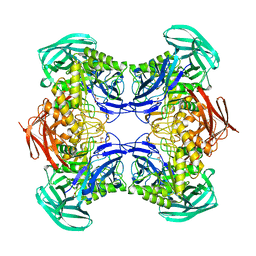 | |
6X8O
 
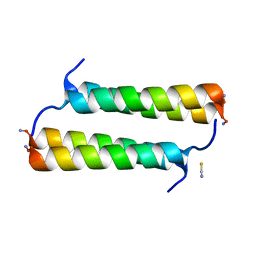 | | BimBH3 peptide tetramer | | Descriptor: | Bcl-2-like protein 11, THIOCYANATE ION | | Authors: | Robin, A.Y, Westphal, D, Uson, I, Czabotar, P.E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-09-09 | | Last modified: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Biophysical Characterization of Pro-apoptotic BimBH3 Peptides Reveals an Unexpected Capacity for Self-Association.
Structure, 29, 2021
|
|
5FPT
 
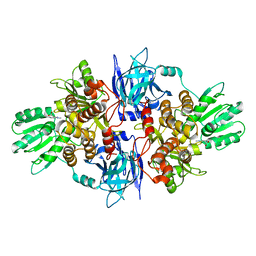 | | Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site. | | Descriptor: | (1-methyl-1H-indol-3-yl)acetic acid, HEPATITIS C VIRUS FULL-LENGTH NS3 COMPLEX | | Authors: | Jhoti, H, Ludlow, R.F, Saini, H.K, Tickle, I.J, Verdonk, M, Pathuri, P, Williams, P.A. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8T81
 
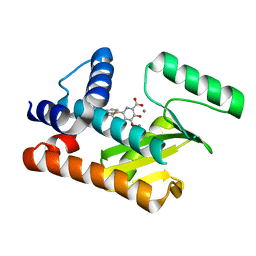 | |
6RHK
 
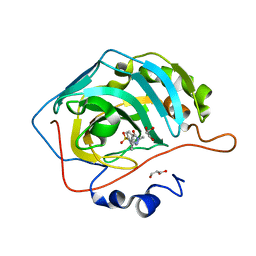 | | The crystal structure of human carbonic anhydrase II in complex with 4-(3-benzylimidazolidine-1-carbonyl)benzenesulfonamide | | Descriptor: | 4-[3-(phenylmethyl)imidazolidin-1-yl]carbonylbenzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ferraroni, M, Angeli, A, Supuran, C.T. | | Deposit date: | 2019-04-22 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Sulfonamides incorporating piperazine bioisosteres as potent human carbonic anhydrase I, II, IV and IX inhibitors.
Bioorg.Chem., 91, 2019
|
|
6DRF
 
 | | Structure of human Retinal Degeneration 3(RD3) Protein | | Descriptor: | Protein RD3 | | Authors: | Yu, Q, Lim, S, Peshenko, I, Cudia, D, Dizhoor, A.M, Ames, J.B. | | Deposit date: | 2018-06-11 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Retinal degeneration 3 (RD3) protein, a retinal guanylyl cyclase regulator, forms a monomeric and elongated four-helix bundle.
J. Biol. Chem., 294, 2019
|
|
8J6D
 
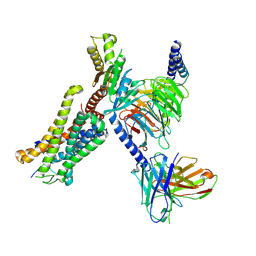 | | Structure of EP141-C3aR-Go complex | | Descriptor: | Antibody fragment - ScFv16, C3a anaphylatoxin chemotactic receptor, EP141 peptide agonist, ... | | Authors: | Yadav, M.K, Yadav, R, Maharana, J, Sarma, P, Banerjee, R, Shukla, A.K, Gati, C. | | Deposit date: | 2023-04-25 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors.
Cell, 186, 2023
|
|
5WDA
 
 | | Structure of the PulG pseudopilus | | Descriptor: | CALCIUM ION, General secretion pathway protein G | | Authors: | Lopez-Castilla, A, Thomassin, J.L, Bardiaux, B, Zheng, W, Nivaskumar, M, Yu, X, Nilges, M, Egelman, E.H, Izadi-Pruneyre, N, Francetic, O. | | Deposit date: | 2017-07-04 | | Release date: | 2017-10-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structure of the calcium-dependent type 2 secretion pseudopilus.
Nat Microbiol, 2, 2017
|
|
8TOL
 
 | |
