8SP6
 
 | | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST | | Descriptor: | (1s,4s)-bicyclo[2.2.1]heptane, (5R0)FAL(MLZ)(5R5), Chromodomain Y-like protein 2, ... | | Authors: | Song, X, Beldar, S, Dong, A, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | COMPLEX STRUCTURE OF CDYL2 WITH AN ANTAGONIST
To be published
|
|
7ASJ
 
 | |
5FQ7
 
 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | BT_2261, BT_2262, BT_2263, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
6PST
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5b) with TraR and mutant rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6VTV
 
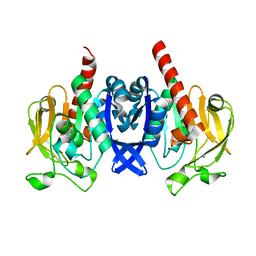 | | Crystal structure of PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase from E. coli | | Descriptor: | Gamma-glutamyl-gamma-aminobutyrate hydrolase PuuD, MANGANESE (II) ION | | Authors: | Stogios, P.J, EVDOKIMOVA, E, DI LEO, R, SAVCHENKO, A, JOACHIMIAK, A, SATCHELL, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-02-13 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | PuuD gamma-glutamyl-gamma-aminobutyrate hydrolase
To Be Published
|
|
8B3P
 
 | |
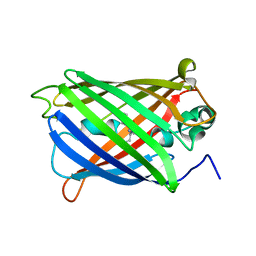
6VIO
 
 | |
8B3O
 
 | |
6PSV
 
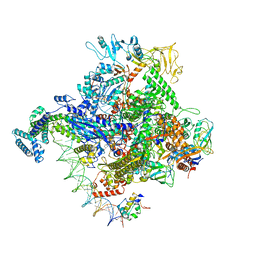 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TpreRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PTT
 
 | | Soluble model of Arabidopsis thaliana CuA (Tt3LAt) | | Descriptor: | Cytochrome c oxidase subunit 2, DINUCLEAR COPPER ION | | Authors: | Lisa, M.N, Giannini, E, Llases, M.E, Alzari, P.M, Vila, A.J. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Unexpected electron spin density on the axial methionine ligand in CuAsuggests its involvement in electron pathways.
Chem.Commun.(Camb.), 56, 2020
|
|
4YD7
 
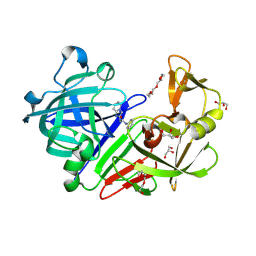 | | Endothiapepsin in complex with fragment 255 | | Descriptor: | 2-(imidazo[1,2-a]pyridin-2-yl)-N-phenylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5J5O
 
 | | Translation initiation factor 4E in complex with m7GppppG mRNA 5' cap analog | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]oxy}phosphoryl]-7-methylguanosine, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Rydzik, A.M, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-04-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.867 Å) | | Cite: | mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation.
Nucleic Acids Res., 45, 2017
|
|
6F2X
 
 | | Structural characterization of the Mycobacterium tuberculosis Protein Tyrosine Kinase A (PtkA) | | Descriptor: | Protein Tyrosine Kinase A | | Authors: | Niesteruk, A, Jonker, H.R.A, Sreeramulu, S, Richter, C, Hutchison, M, Linhard, V, Schwalbe, H. | | Deposit date: | 2017-11-27 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The domain architecture of PtkA, the first tyrosine kinase fromMycobacterium tuberculosis, differs from the conventional kinase architecture.
J. Biol. Chem., 293, 2018
|
|
6VY1
 
 | | Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii | | Descriptor: | Prefoldin subunit alpha 2 | | Authors: | Wang, F, Chen, Y.X, Ing, N.L, Hochbaum, A.I, Clark, D.S, Glover, D.J, Egelman, E.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Determination of a Filamentous Chaperone to Fabricate Electronically Conductive Metalloprotein Nanowires.
Acs Nano, 14, 2020
|
|
8RXR
 
 | | Crystal structure of VPS34 in complex with inhibitor SB02024 | | Descriptor: | 4-[(3R)-3-methylmorpholin-4-yl]-2-[(2R)-2-(trifluoromethyl)piperidin-1-yl]-3H-pyridin-6-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Tresaugues, L, Yu, Y, Bogdan, M, Parpal, S, Silvander, C, Lindstrom, J, Simeon, J, Timson, M.J, Al-Hashimi, H, Smith, B.D, Flynn, D.L, Viklund, J, Martinsson, J, De Milito, A, Andersson, M. | | Deposit date: | 2024-02-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Combining VPS34 inhibitors with STING agonists enhances type I interferon signaling and anti-tumor efficacy.
Mol Oncol, 18, 2024
|
|
4YQR
 
 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)(4-methylpiperazin-1-yl)methanone, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
6YQI
 
 | | Crystal structure of cAMP-dependent Protein Kinase (PKA) in complex with long-chain Fasudil-derivative N-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide (soaked) | | Descriptor: | cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha, ~{N}-[2-(propylamino)ethyl]isoquinoline-5-sulfonamide | | Authors: | Oebbeke, M, Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2020-04-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Two Methods, One Goal: Structural Differences between Cocrystallization and Crystal Soaking to Discover Ligand Binding Poses.
Chemmedchem, 16, 2021
|
|
6BH9
 
 | | Caspase-3 Mutant - T152A | | Descriptor: | Ac-Asp-Glu-Val-Asp-CMK, CHLORIDE ION, Caspase-3, ... | | Authors: | Thomas, M.E, Grinshpon, R, Swartz, P.D, Clark, A.C. | | Deposit date: | 2017-10-30 | | Release date: | 2018-02-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Modifications to a common phosphorylation network provide individualized control in caspases.
J. Biol. Chem., 293, 2018
|
|
5JAG
 
 | | LeuT T354H mutant in the outward-oriented, Na+-free Return State | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
6PYI
 
 | |
7ZN4
 
 | | Tail tip of siphophage T5 : bent fibre after interaction with its bacterial receptor FhuA | | Descriptor: | Probable baseplate hub protein, Probable central straight fiber | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-20 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
3LZL
 
 | | Crystal Structure Analysis of the as-solated P19 protein from Campylobacter jejuni at 1.45 A at pH 9.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
8UTW
 
 | | KIF1A[1-393] P305L mutant APO in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF1A, ... | | Authors: | Benoit, M.P.M.H, Rao, L, Asenjo, A.B, Gennerich, A, Sosa, H. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM unveils kinesin KIF1A's processivity mechanism and the impact of its pathogenic variant P305L.
Nat Commun, 15, 2024
|
|
5HOI
 
 | |
4YUR
 
 | | Crystal Structure of Plk4 Kinase Domain Bound to Centrinone | | Descriptor: | 2-({2-fluoro-4-[(2-fluoro-3-nitrobenzyl)sulfonyl]phenyl}sulfanyl)-5-methoxy-N-(3-methyl-1H-pyrazol-5-yl)-6-(morpholin-4-yl)pyrimidin-4-amine, Serine/threonine-protein kinase PLK4 | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4.
Science, 348, 2015
|
|
