6V93
 
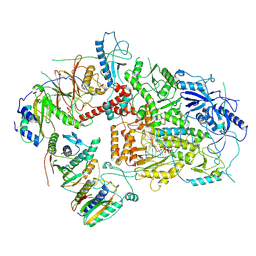 | | Structure of DNA Polymerase Zeta/DNA/dNTP Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA, ... | | Authors: | Malik, R, Kopylov, M, Jain, R, Ubarrextena-Belandia, I, Aggarwal, A.K. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6HN3
 
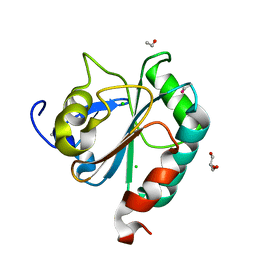 | | wildtype form (apo) of human GPX4 with Se-Cys46 | | Descriptor: | CHLORIDE ION, ETHANOL, GLYCEROL, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Hoffmann, J, Schnirch, L, Eaton, J.K, Badock, V, Gradl, S. | | Deposit date: | 2018-09-13 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4GXS
 
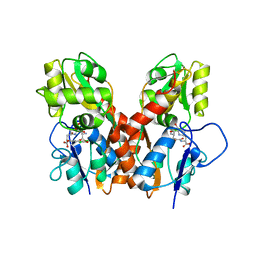 | | Ligand binding domain of GluA2 (AMPA/glutamate receptor) bound to (-)-kaitocephalin | | Descriptor: | (5R)-2-[(1S,2R)-2-amino-2-carboxy-1-hydroxyethyl]-5-{(2S)-2-carboxy-2-[(3,5-dichloro-4-hydroxybenzoyl)amino]ethyl}-L-proline, Glutamate receptor 2, ZINC ION | | Authors: | Ahmed, A.H, Oswald, R.E. | | Deposit date: | 2012-09-04 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9634 Å) | | Cite: | The structure of (-)-kaitocephalin bound to the ligand binding domain of the (S)-alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA)/glutamate receptor, GluA2.
J.Biol.Chem., 287, 2012
|
|
5TNO
 
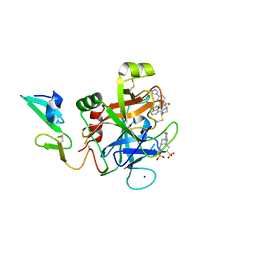 | | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Coagulation factor IX, SODIUM ION, ... | | Authors: | Sakurada, I, Endo, T, Hikita, K, Hirabayashi, T, Hosaka, Y, Kato, Y, Maeda, Y, Matsumoto, S, Mizuno, T, Nagasue, H, Nishimura, T, Shimada, S, Shinozaki, M, Taguchi, K, Takeuchi, K, Yokoyama, T, Hruza, A, Reichert, P, Zhang, T, Wood, H.B, Nakao, K, Furusako, S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel aminobenzisoxazole derivatives as orally available factor IXa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5H9A
 
 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
4R7U
 
 | | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Nocek, B, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin
To be Published
|
|
4ZD6
 
 | | Halohydrin hydrogen-halide-lyase, HheB | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Halohydrin epoxidase B, ... | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2015-04-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of halohydrin hydrogen-halide-lyases from Corynebacterium sp. N-1074
Proteins, 83, 2015
|
|
2WXK
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK666. | | Descriptor: | 3-(2-amino-1,3-benzothiazol-6-yl)-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
5H83
 
 | | HETEROYOHIMBINE SYNTHASE HYS FROM CATHARANTHUS ROSEUS - APO FORM | | Descriptor: | ZINC ION, heteroyohimbine synthase HYS | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-23 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
3FN8
 
 | |
6T0P
 
 | | Cationic Trypsin in Complex with a D-Phe-Pro-2-aminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(2-azanylpyridin-4-yl)methyl]pyrrolidine-2-carboxamide, CALCIUM ION, Cationic Trypsin, ... | | Authors: | Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | Authors: | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-07 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6HRS
 
 | | Structure of the TRPML2 ELD at pH 4.5 | | Descriptor: | GLYCEROL, Mucolipin-2 | | Authors: | Bader, N, Viet, K.K, Wagner, A, Hellmich, U.A, Schindelin, H. | | Deposit date: | 2018-09-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain.
Structure, 27, 2019
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | Descriptor: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-29 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
6NR3
 
 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
4ZG6
 
 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | 4-{(Z)-2-[6-chloro-1-(4-fluorobenzyl)-1H-indol-3-yl]-1-cyanoethenyl}benzoic acid, CALCIUM ION, Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
3OU9
 
 | |
7V9K
 
 | | Telomeric tetranucleosome | | Descriptor: | DNA (539-mer), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Soman, A. | | Deposit date: | 2021-08-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Columnar structure of human telomeric chromatin.
Nature, 609, 2022
|
|
7JTK
 
 | | Radial spoke 1 isolated from Chlamydomonas reinhardtii | | Descriptor: | Cytochrome b5 heme-binding domain-containing protein, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6STG
 
 | | Human Rab8a phosphorylated at Ser111 in complex with GPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vieweg, S, Mulholland, K, Braeuning, B, Kachariya, N, Lai, Y, Toth, R, Sattler, M, Groll, M, Itzen, A, Muqit, M.M.K. | | Deposit date: | 2019-09-10 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PINK1-dependent phosphorylation of Serine111 within the SF3 motif of Rab GTPases impairs effector interactions and LRRK2-mediated phosphorylation at Threonine72.
Biochem.J., 477, 2020
|
|
6UZQ
 
 | |
4EPV
 
 | | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-mediated Activation | | Descriptor: | 2-(1H-indol-3-ylmethyl)-1H-imidazo[4,5-c]pyridine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sun, Q, Burke, J.P, Phan, J, Burns, M.C, Olejniczak, E.T, Waterson, A.G, Lee, T, Rossanese, O.W, Fesik, S.W. | | Deposit date: | 2012-04-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of Small Molecules that Bind to K-Ras and Inhibit Sos-Mediated Activation.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7CKB
 
 | | Simplified Alpha-Carboxysome, T=3 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
7CKC
 
 | | Simplified Alpha-Carboxysome, T=4 | | Descriptor: | Major carboxysome shell protein 1A, Unidentified carboxysome polypeptide | | Authors: | Tan, Y.Q, Ali, S, Xue, B, Robinson, R.C, Narita, A, Yew, W.S. | | Deposit date: | 2020-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of a Minimal alpha-Carboxysome-Derived Shell and Its Utility in Enzyme Stabilization.
Biomacromolecules, 22, 2021
|
|
5JOQ
 
 | | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, CITRIC ACID, Lmo2184 protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-02 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e
To Be Published
|
|
