6HXW
 
 | | structure of human CD73 in complex with antibody IPH53 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-nucleotidase, IPH53 heavy chain, ... | | Authors: | Roussel, A, Amigues, B. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies.
Cell Rep, 27, 2019
|
|
4TVT
 
 | | New ligand for thaumatin discovered using acoustic high throughput screening | | Descriptor: | ASCORBIC ACID, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Teplitsky, E, Joshi, K, Ericson, D.L, Scalia, A, Mullen, J.D, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
8F37
 
 | |
7SIZ
 
 | | C-type inactivation in a voltage gated K+ channel | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Voltage gated potassium channel Kv1.2-Kv2.1, ... | | Authors: | Reddi, R, Riederer, E.A, Matulef, K, Whorton, M.R, Valiyaveetil, F.I. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for C-type inactivation in a Shaker family voltage-gated K + channel.
Sci Adv, 8, 2022
|
|
6PSO
 
 | | Crystal structure of PsS1_19B C77S in complex with iota-neocarratetraose | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-13 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
6HGE
 
 | |
4TW3
 
 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
6QJN
 
 | |
6U10
 
 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, L-CAPTOPRIL, ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia in the complex with the inhibitor captopril.
To Be Published
|
|
4UAF
 
 | |
5DAR
 
 | |
7S6I
 
 | | SARS-CoV-2-6P-Mut2 S protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6PT6
 
 | | Crystal structure of PsS1_NC C84S in complex with i-neocarratetraose | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, exo-2S-iota carrageenan S1 sulfatase | | Authors: | Hettle, A.G, Boraston, A.B. | | Deposit date: | 2019-07-15 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the kappa / iota-carrageenan metabolism pathway of some marinePseudoalteromonasspecies.
Commun Biol, 2, 2019
|
|
7TZV
 
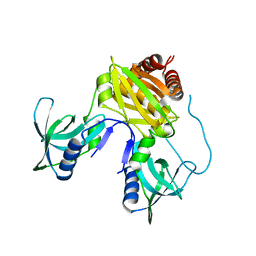 | | Structure of DriD C-domain bound to 9mer ssDNA | | Descriptor: | DNA (5'-D(*TP*AP*GP*TP*CP*TP*AP*CP*T)-3'), WYL domain-containing protein | | Authors: | Schumacher, M.A, Laub, M. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ssDNA is an allosteric regulator of the C. crescentus SOS-independent DNA damage response transcription activator, DriD.
Genes Dev., 36, 2022
|
|
6FXC
 
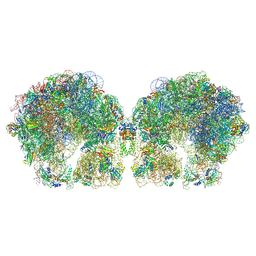 | | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Kidmose, R, Amunts, A, Yonath, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-03-21 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (6.76 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
7S6J
 
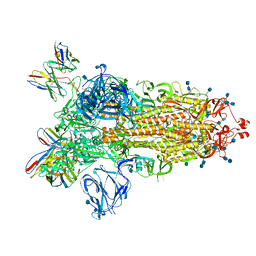 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4UBC
 
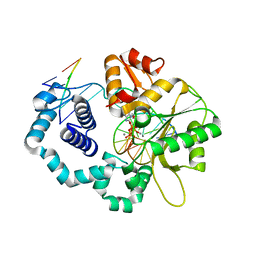 | | DNA polymerase beta substrate complex with a templating cytosine and incoming 8-oxodGTP, 0 s | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*CP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Wilson, S.H, Beard, W.A. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Nature, 517, 2015
|
|
8KB2
 
 | | Crystal Structure of M- and C-Domains of the shaft pilin LrpA from Ligilactobacillus ruminis - iodide derivative | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prajapati, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2023-08-03 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure of the N-terminal domain of the backbone pilin LrpA reveals a new closure-and-twist motion for assembling dynamic pili in Ligilactobacillus ruminis.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6G3K
 
 | | MHC A02 Allele presenting MTSAIGILPV | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2018-03-26 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Peptide Super-Agonist Enhances T-Cell Responses to Melanoma.
Front Immunol, 10, 2019
|
|
7S6L
 
 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5JRO
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form | | Descriptor: | FMN-dependent NADH-azoreductase, GLYCEROL | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-06 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | The crystal structure of azoreductase from Yersinia pestis CO92 in its Apo form
To Be Published
|
|
6G44
 
 | | Crystal structure of mavirus major capsid protein lacking the C-terminal domain | | Descriptor: | GLYCEROL, Putative major capsid protein, SULFATE ION | | Authors: | Born, D, Reuter, L, Meinhart, A, Reinstein, J. | | Deposit date: | 2018-03-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capsid protein structure, self-assembly, and processing reveal morphogenesis of the marine virophage mavirus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7S6K
 
 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6PXL
 
 | | 3.74 Angstroms resolution structure of HlsU with an axial-channel plug | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, MAGNESIUM ION, ... | | Authors: | Baytshtok, V, Grant, R.A, Sauer, R.T. | | Deposit date: | 2019-07-26 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.741 Å) | | Cite: | Heat activates the AAA+ HslUV protease by melting an axial autoinhibitory plug.
Cell Rep, 34, 2021
|
|
8ERM
 
 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellin, GLYCEROL, SULFATE ION | | Authors: | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
