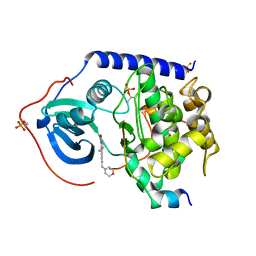
5N23
 
 | |
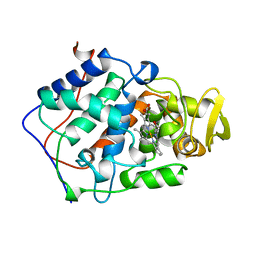
1BEJ
 
 | |
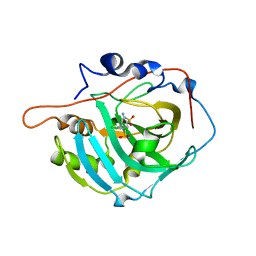
7PLF
 
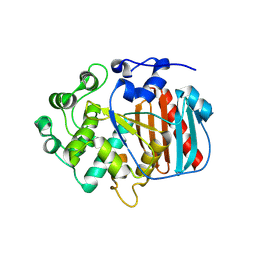
 | | Human Carbonic Anhydrase I in complex with clorsulon | | Descriptor: | 4-azanyl-6-[1,2,2-tris(chloranyl)ethenyl]benzene-1,3-disulfonamide, Carbonic anhydrase 1, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.461 Å) | | Cite: | Inhibition of Schistosoma mansoni carbonic anhydrase by the antiparasitic drug clorsulon: X-ray crystallographic and in vitro studies.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8RLJ
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | Class C beta-lactamase-related serine hydrolase | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
6Y21
 
 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with L-Cystathionine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Cystathionine Beta-synthase from Toxoplasma gondii with PLP-Cystathionine
To Be Published
|
|
6MI1
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE VARIANT PLANT EXOHYDROLASE ARG158ALA-GLU161ALA IN COMPLEX WITH METHYL 6-THIO-BETA-GENTIOBIOSIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-D-glucan exohydrolase isoenzyme ExoI, GLYCEROL, ... | | Authors: | Streltsov, V.A, Luang, S, Hrmova, M. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of processive catalysis by an exo-hydrolase with a pocket-shaped active site.
Nat Commun, 10, 2019
|
|
4RJ1
 
 | | Structural variations and solvent structure of UGGGGU quadruplexes stabilized by Sr2+ ions | | Descriptor: | CALCIUM ION, RNA (5'-R(*UP*GP*GP*GP*GP*U)-3'), SODIUM ION, ... | | Authors: | Fyfe, A.C, Dunten, P.W, Scott, W.G. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Structural Variations and Solvent Structure of r(UGGGGU) Quadruplexes Stabilized by Sr(2+) Ions.
J.Mol.Biol., 427, 2015
|
|
8RLK
 
 | | Structure of the apo form of PIB-1 in an Orthorombic space group | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Class C beta-lactamase-related serine hydrolase, MAGNESIUM ION, ... | | Authors: | Medrano, F.J, Romero, A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new type of Class C beta-lactamases defined by PIB-1. A metal-dependent carbapenem-hydrolyzing beta-lactamase, from Pseudomonas aeruginosa: Structural and functional analysis.
Int.J.Biol.Macromol., 277, 2024
|
|
7L3N
 
 | | SARS-CoV 2 Spike Protein bound to LY-CoV555 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV555 Fab heavy chain, ... | | Authors: | Goldsmith, J.A, McLellan, J.S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection.
Biorxiv, 2020
|
|
8RVN
 
 | | Nipah virus (NiV) fusion protein in complex with neutralizing Fab92 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab92 heavy chain, Fab92 light chain, ... | | Authors: | Avanzato, V.A, Stass, R, Duyvesteyn, H.M.E, Bowden, T.A. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A monoclonal antibody targeting the Nipah virus fusion glycoprotein apex imparts protection from disease.
J.Virol., 2024
|
|
6Q85
 
 | | Structure of Fucosylated D-antimicrobial peptide SB11 in complex with the Fucose-binding lectin PA-IIL at 1.990 Angstrom resolution | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, AMINO GROUP, CALCIUM ION, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2018-12-14 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | X-ray Crystal Structures of Short Antimicrobial Peptides as Pseudomonas aeruginosa Lectin B Complexes.
Acs Chem.Biol., 14, 2019
|
|
8RU3
 
 | | Crystal structure of beta-catenin in complex with alpha-helical peptide inhibitor | | Descriptor: | Axin-1, CHLORIDE ION, Catenin beta-1 | | Authors: | Yeste Vazquez, A, Klintrot, C.I.R, Grossmann, T.N, Hennig, S. | | Deposit date: | 2024-01-30 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structure-Based Design of Bicyclic Helical Peptides That Target the Oncogene beta-Catenin.
Angew.Chem.Int.Ed.Engl., 2024
|
|
5GG8
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP, 8-oxo-dGMP and pyrophosphate (II) | | Descriptor: | 8-OXO-2'-DEOXY-GUANOSINE-5'-MONOPHOSPHATE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6SIS
 
 | | Crystal structure of macrocyclic PROTAC 1 in complex with the second bromodomain of human Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Hughes, S.J, Testa, A, Ciulli, A. | | Deposit date: | 2019-08-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Based Design of a Macrocyclic PROTAC.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5GG9
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-GTP, 8-oxo-GMP and pyrophosphate | | Descriptor: | 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, Hydrolase, NUDIX family protein, ... | | Authors: | Arif, S.M, Patil, A.G, Varshney, U, Vijayan, M. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural studies of Mycobacterium smegmatis MutT1, a sanitization enzyme with unusual modes of association
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4YLU
 
 | | X-ray structure of MERS-CoV nsp5 protease bound with a non-covalent inhibitor | | Descriptor: | ACETATE ION, N-{4-[(1H-benzotriazol-1-ylacetyl)(thiophen-3-ylmethyl)amino]phenyl}propanamide, ORF1a protein | | Authors: | Tomar, S, Mesecar, A.D. | | Deposit date: | 2015-03-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro): IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS.
J.Biol.Chem., 290, 2015
|
|
6F7L
 
 | | Crystal structure of LkcE R326Q mutant in complex with its substrate | | Descriptor: | ACETATE ION, Amine oxidase LkcE, CALCIUM ION, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
6YRF
 
 | | Vip3Bc1 tetramer | | Descriptor: | Vegetative insecticidal protein | | Authors: | Thompson, R.F, Byrne, M.J, Iadanza, M.I, Arribas Perez, M, Maskell, D.P, George, R.M, Hesketh, E.L, Beales, P.A, Zack, M.D, Berry, C. | | Deposit date: | 2020-04-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of an insecticidal Bt toxin reveal its mechanism of action on the membrane.
Nat Commun, 12, 2021
|
|
7ZB9
 
 | | Crystal structure of CYP124 in complex with inhibitor carbethoxyhexyl imidazole in the absence of glycerol (NoCryo) | | Descriptor: | CHLORIDE ION, CYP124 in complex with inhibitor carbethoxyhexyl imidazole, MAGNESIUM ION, ... | | Authors: | Bukhdruker, S, Varaksa, T, Marin, E, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the effects of glycerol on ligand binding to cytochrome P450.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
3M41
 
 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
7UV7
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold duplex (21mer) containing the cognate REPE54 sequence and an insert duplex (10mer) with guest TAMRA-labelled thymine and T-A rich sequence. | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*TP*T)-3'), DNA (5'-D(*GP*T(TAMRA)P*AP*AP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
6SLM
 
 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | Descriptor: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | Authors: | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | Deposit date: | 2019-08-20 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
4RGM
 
 | | Structure of Staphylococcal Enterotoxin B bound to the neutralizing antibody 20B1 | | Descriptor: | 20B1 heavy chain, 20B1 light chain, Enterotoxin type B | | Authors: | Franklin, M.C, Dutta, K, Varshney, A.K, Goger, M.J, Fries, B.C. | | Deposit date: | 2014-09-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Mechanisms mediating enhanced neutralization efficacy of staphylococcal enterotoxin B by combinations of monoclonal antibodies.
J.Biol.Chem., 290, 2015
|
|
6MLV
 
 | | Crystal structure of the periplasmic Lysine-, Arginine-, Ornithine-binding protein (LAO) Y14A mutant from Salmonella typhimurium | | Descriptor: | Lysine/arginine/ornithine-binding periplasmic protein | | Authors: | Romero-Romero, S, Vergara, R, Espinoza-Perez, G, Rodriguez-Romero, A. | | Deposit date: | 2018-09-28 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.082 Å) | | Cite: | The interplay of protein-ligand and water-mediated interactions shape affinity and selectivity in the LAO binding protein.
Febs J., 287, 2020
|
|
