4X0E
 
 | |
6EEB
 
 | | Calmodulin in complex with malbrancheamide | | Descriptor: | (5aS,12aS,13aS)-8,9-dichloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CALCIUM ION, Calmodulin-1, ... | | Authors: | Beyett, T.S, Fraley, A.E, Tesmer, J.J.G. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Perturbation of the interactions of calmodulin with GRK5 using a natural product chemical probe.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6VRM
 
 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
6EHN
 
 | | Structural insight into a promiscuous CE15 esterase from the marine bacterial metagenome | | Descriptor: | Carbohydrate esterase MZ0003, GLYCEROL | | Authors: | Helland, R, De Santi, C, Gani, O, Williamson, A.K. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into a CE15 esterase from the marine bacterial metagenome.
Sci Rep, 7, 2017
|
|
7S88
 
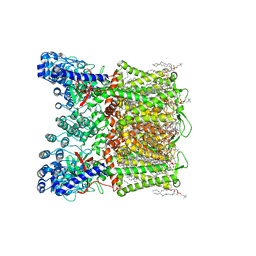 | | Open apo-state cryo-EM structure of human TRPV6 in glyco-diosgenin detergent | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Sobolevsky, A.I. | | Deposit date: | 2021-09-17 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural mechanisms of TRPV6 inhibition by ruthenium red and econazole.
Nat Commun, 12, 2021
|
|
8AU1
 
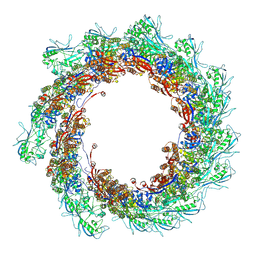 | | Jumbo Phage phi-kp24 tail outer sheath | | Descriptor: | Putative tail sheath protein | | Authors: | Ouyang, R, Briegel, A. | | Deposit date: | 2022-08-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution reconstruction of a Jumbo-bacteriophage infecting capsulated bacteria using hyperbranched tail fibers.
Nat Commun, 13, 2022
|
|
6EC6
 
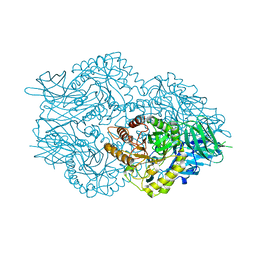 | | Ruminococcus gnavus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Redinbo, M.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
5JI2
 
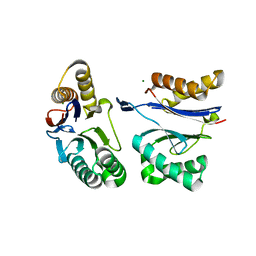 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
6RMT
 
 | |
6YIF
 
 | | Structure of Chromosomal Passenger Complex (CPC) bound to phosphorylated Histone 3 peptide at 1.8 A. | | Descriptor: | Baculoviral IAP repeat-containing protein 5, Borealin, Inner centromere protein, ... | | Authors: | Serena, M, Elliott, P.R, Barr, F.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Molecular basis of MKLP2-dependent Aurora B transport from chromatin to the anaphase central spindle.
J.Cell Biol., 219, 2020
|
|
6S7E
 
 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SHE
 
 | | Cryo-EM structure of human GPR158 | | Descriptor: | (2S)-1-{[(S)-hydroxy{[(1s,2R,3R,4R,5S,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5E,8E,11E,14E)-icosa-5,8,11,14-tetraenoate, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, ... | | Authors: | Patil, D.N, Singh, S, Singh, A.K, Martemyanov, K.A. | | Deposit date: | 2021-10-08 | | Release date: | 2021-12-01 | | Last modified: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of human GPR158 receptor coupled to the RGS7-G beta 5 signaling complex.
Science, 375, 2022
|
|
5LO7
 
 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Fimbrial protein MyfA,Fimbrial protein MyfA | | Authors: | Pakharukova, N.A, Roy, S, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-08 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
6S7U
 
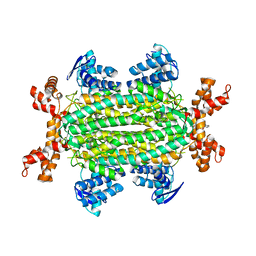 | | Fumarate hydratase of Mycobacterium tuberculosis in complex with formate and allosteric modulator N-(5-(Azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(1H-indol-3-yl)acetamide | | Descriptor: | FORMIC ACID, Fumarate hydratase class II, MAGNESIUM ION, ... | | Authors: | Whitehouse, A.J, Libardo, M.D, Kasbekar, M, Brear, P, Fischer, G, Thomas, C.J, Barry, C.E, Boshoff, H.I, Coyne, A.G, Abell, C. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Targeting of Fumarate Hydratase fromMycobacterium tuberculosisUsing Allosteric Inhibitors with a Dimeric-Binding Mode.
J.Med.Chem., 62, 2019
|
|
8IK8
 
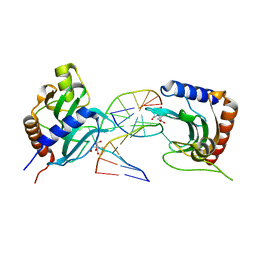 | |
5JJN
 
 | | Structure of the SRII/HtrII(G83F) Complex in P212121 space group ("V" shape) | | Descriptor: | EICOSANE, RETINAL, Sensory rhodopsin II transducer, ... | | Authors: | Ishchenko, A, Round, E, Borshchevskiy, V, Grudinin, S, Gushchin, I, Klare, J, Remeeva, A, Polovinkin, V, Utrobin, P, Balandin, T, Engelhard, M, Bueldt, G, Gordeliy, V. | | Deposit date: | 2016-04-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | New Insights on Signal Propagation by Sensory Rhodopsin II/Transducer Complex.
Sci Rep, 7, 2017
|
|
5FQE
 
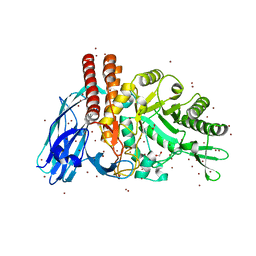 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BETA-N-ACETYLGALACTOSAMINIDASE, BROMIDE ION, ... | | Authors: | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
6VY0
 
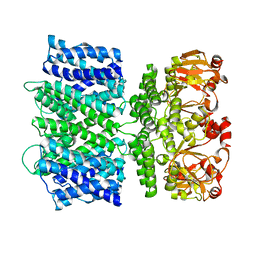 | |
5TCU
 
 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
5LRB
 
 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
7Z5E
 
 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7U8J
 
 | |
6NHW
 
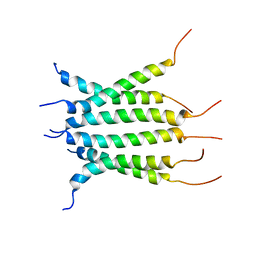 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
7Z5F
 
 | | VP2-only capsid of MVM D263A mutant | | Descriptor: | Capsid protein VP1 | | Authors: | Luque, D, Ortega-Esteban, A, Valbuena, A, Vilas, J.L, Rodriguez-Huete, A, Mateu, M.G, Caston, J.R. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Equilibrium Dynamics of a Biomolecular Complex Analyzed at Single-amino Acid Resolution by Cryo-electron Microscopy.
J.Mol.Biol., 435, 2023
|
|
7TYG
 
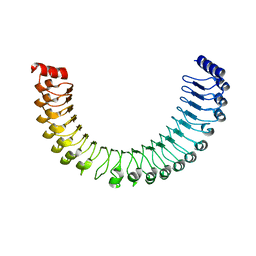 | |
