8DMO
 
 | | Structure of open, inward-facing MsbA from E. coli | | Descriptor: | ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
5MI1
 
 | | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 9-th structure of the series with total exposition time 243 min. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2016-11-27 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: insights into the reaction mechanism.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6SRP
 
 | |
8DMM
 
 | | Structure of the vanadate-trapped MsbA bound to KDL | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, ADP ORTHOVANADATE, ATP-binding transport protein MsbA | | Authors: | Liu, C, Lyu, J, Laganowsky, A.D, Zhao, M. | | Deposit date: | 2022-07-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for lipid and copper regulation of the ABC transporter MsbA.
Nat Commun, 13, 2022
|
|
5MIB
 
 | | The study of the X-ray induced enzymatic reduction of molecular oxygen to water for laccase from Steccherinum murashkinskyi.The 12-th structure of the series with total exposition time 333 min. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2016-11-28 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural study of the X-ray-induced enzymatic reduction of molecular oxygen to water by Steccherinum murashkinskyi laccase: insights into the reaction mechanism.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
6GUZ
 
 | | Ground state structure of Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich at room temperature | | Descriptor: | Archaerhodopsin-3, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Moraes, I, Judge, P.J, Bada Juarez, J.F, Vinals, J, Axford, D, Watts, A. | | Deposit date: | 2018-06-19 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Room temperature structure of the Archaerhodopsin-3 obtained from LCP crystals using a thin-film sandwich
To Be Published
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | Descriptor: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | Authors: | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-05-22 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
6Y76
 
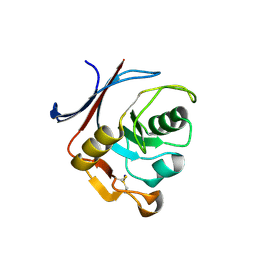 | | AP01 - a redesigned transferrin receptor apical domain | | Descriptor: | SODIUM ION, Transferrin receptor protein 1 | | Authors: | Oberdorfer, G, Berger, S.A, Bjelic, S, Sjostrom, D.J. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Computational backbone design enables soluble engineering of transferrin receptor apical domain.
Proteins, 88, 2020
|
|
5K7C
 
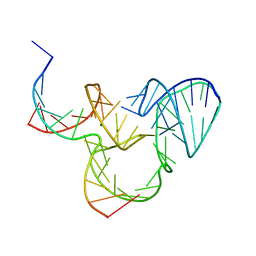 | | The native structure of native pistol ribozyme | | Descriptor: | DNA/RNA 11-MER, MAGNESIUM ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
7ASO
 
 | |
7R5L
 
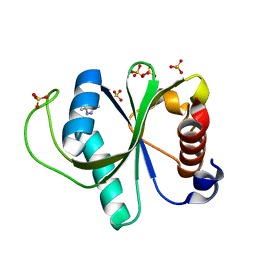 | | Crystal structure of YTHDF2 with compound YLI_DC1_015 | | Descriptor: | 3,6-dimethyl-2~{H}-1,2,4-triazin-5-one, SULFATE ION, YTH domain-containing family protein 2 | | Authors: | Nachawati, R, Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
8V1J
 
 | |
4Y4T
 
 | | Endothiapepsin in complex with fragment 114 | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-5-(pyrrolidin-1-yl)-1H-pyrazole-4-carbonitrile, ACETATE ION, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
8RJY
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3899 (compound 58 in publication) | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-1-[[(2~{S})-1-[[(4-chlorophenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]amino]-3,3-dimethyl-1-oxidanylidene-butan-2-yl]thiophene-2-carboxamide | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
5MB6
 
 | |
7STU
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | BROMIDE ION, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
7L08
 
 | | Cryo-EM structure of the human 55S mitoribosome-RRFmt complex. | | Descriptor: | 12S rRNA, 16S RRNA, 28S ribosomal protein S10, ... | | Authors: | Koripella, R, Agrawal, E.K, Deep, A, Agrawal, R.K. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-12 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Distinct mechanisms of the human mitoribosome recycling and antibiotic resistance.
Nat Commun, 12, 2021
|
|
5MBQ
 
 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
8QVD
 
 | | Deinococcus aerius TR0125 C-glucosyl deglycosidase (CGD), wild type crystal cryoprotected with glycerol | | Descriptor: | CADMIUM ION, DUF6379 domain-containing protein, Xylose isomerase-like TIM barrel domain-containing protein | | Authors: | Furlanetto, V, Kalyani, D.C, Kostelac, A, Haltrich, D, Hallberg, B.M, Divne, C. | | Deposit date: | 2023-10-17 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Characterization of a Gene Cluster Responsible for Deglycosylation of C-glucosyl Flavonoids and Xanthonoids by Deinococcus aerius.
J.Mol.Biol., 436, 2024
|
|
7STT
 
 | | Crystal structure of sulfatase from Pedobacter yulinensis | | Descriptor: | CALCIUM ION, CHLORIDE ION, MALONATE ION, ... | | Authors: | O'Malley, A, Schlachter, C.R, Grimes, L.L, Tomashek, J.J, Lee, A.L, Chruszcz, M. | | Deposit date: | 2021-11-15 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Purification, Characterization, and Structural Studies of a Sulfatase from Pedobacter yulinensis .
Molecules, 27, 2021
|
|
4Y3L
 
 | | Endothiapepsin in complex with fragment 205 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
6NRB
 
 | | hTRiC-hPFD Class2 | | Descriptor: | Prefoldin subunit 1, Prefoldin subunit 2, Prefoldin subunit 3, ... | | Authors: | Gestaut, D.R, Roh, S.H, Ma, B, Pintilie, G, Joachimiak, L.A, Leitner, A, Walzthoeni, T, Aebersold, R, Chiu, W, Frydman, J. | | Deposit date: | 2019-01-23 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | The Chaperonin TRiC/CCT Associates with Prefoldin through a Conserved Electrostatic Interface Essential for Cellular Proteostasis.
Cell, 177, 2019
|
|
8RJV
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the covalent inhibitor GUE-3778 (compound 12 in publication) | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-1-[[(3-chloranyl-2-fluoranyl-phenyl)methyl-(iminomethyl)amino]-methyl-amino]-1-oxidanylidene-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Strater, N, Claff, T, Sylvester, K, Mueller, C.E, Guetschow, M, Useini, A. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Macrocyclic Azapeptide Nitriles: Structure-Based Discovery of Potent SARS-CoV-2 Main Protease Inhibitors as Antiviral Drugs.
J.Med.Chem., 67, 2024
|
|
6SW8
 
 | | Crystal structure of the NS1 (H7N1) RNA-binding domain | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Non-structural protein 1 | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
