6OCT
 
 | | Crystal structure of human KCTD16 T1 domain | | Descriptor: | BTB/POZ domain-containing protein KCTD16 | | Authors: | Zuo, H, Glaaser, I, Zhao, Y, Kurinov, I, Mosyak, L, Wang, H, Liu, J, Park, J, Frangaj, A, Sturchler, E, Zhou, M, McDonald, P, Geng, Y, Slesinger, P.A, Fan, Q.R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for auxiliary subunit KCTD16 regulation of the GABABreceptor.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6U55
 
 | | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire E (ZE) Complexed with Sudan ebolavirus Nucleoprotein C-terminal Domain 634-738 | | Descriptor: | Anti-Zaire ebolavirus Nucleoprotein Single Domain Antibody Zaire E (ZE), Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
6MF4
 
 | |
6MFB
 
 | | Crystal structure of 3-hydroxykynurenine transaminase from Aedes aegypti | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Maciel, L.G, Oliveira, A.A, Romao, T.P, Silva Filha, M.H.N.L, dos Anjos, J.V, Soares, T.A, Guido, R.V.C. | | Deposit date: | 2018-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of 3-hydroxykynurenine transaminase from Aedes aegypti
To Be Published
|
|
8BW0
 
 | | Structure of CEACAM5 A3-B3 domain in Complex with Tusamitamab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 5, ... | | Authors: | Kumar, A, Bertrand, T, Rapisarda, C, Rak, A. | | Deposit date: | 2022-12-06 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural insights into epitope-paratope interactions of monoclonal antibody targeting CEACAM5-expressing tumors
Res Sq, 2023
|
|
6XN0
 
 | | Crystal structure of GH43_1 enzyme from Xanthomonas citri | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Morais, M.A.B, Tonoli, C.C.C, Santos, C.R, Murakami, M.T. | | Deposit date: | 2020-07-02 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Two distinct catalytic pathways for GH43 xylanolytic enzymes unveiled by X-ray and QM/MM simulations.
Nat Commun, 12, 2021
|
|
5LNH
 
 | | Structure of full length Unliganded CodY from Bacillus subtilis | | Descriptor: | GTP-sensing transcriptional pleiotropic repressor CodY, SULFATE ION | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2016-08-04 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Branched-chain Amino Acid and GTP-sensing Global Regulator, CodY, from Bacillus subtilis.
J. Biol. Chem., 292, 2017
|
|
8E2X
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3C
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
6OG1
 
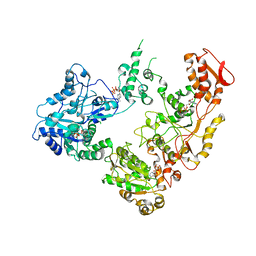 | | Focus classification structure of the hyperactive ClpB mutant K476C, bound to casein, pre-state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hyperactive disaggregase ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Rizo, A.R, Lin, J.-B, Gates, S.N, Tse, E, Bart, S.M, Castellano, L.M, Dimaio, F, Shorter, J, Southworth, D.R. | | Deposit date: | 2019-04-01 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for substrate gripping and translocation by the ClpB AAA+ disaggregase.
Nat Commun, 10, 2019
|
|
6UIU
 
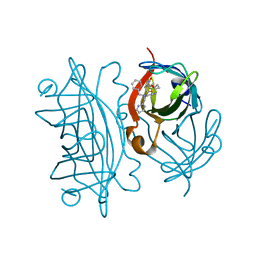 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | N-(2-{bis[(pyridin-2-yl)methyl]amino}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide, Streptavidin | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
7S8H
 
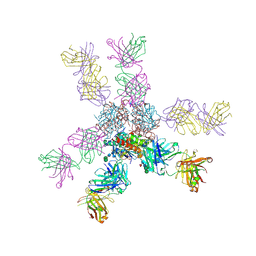 | |
8GM7
 
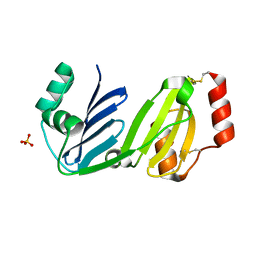 | |
7N7Z
 
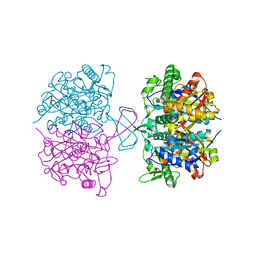 | |
5IGY
 
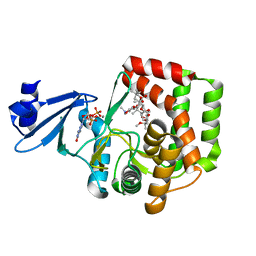 | |
8E1E
 
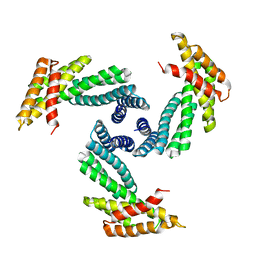 | |
6UKJ
 
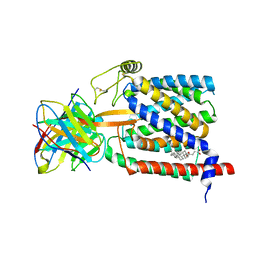 | | Single-Particle Cryo-EM Structure of Plasmodium falciparum Chloroquine Resistance Transporter (PfCRT) 7G8 Isoform | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Chloroquine resistance transporter, Fab Heavy Chain, ... | | Authors: | Kim, J, Tan, Y.Z, Wicht, K.J, Erramilli, S.K, Dhingra, S.K, Okombo, J, Vendome, J, Hagenah, L.M, Giacometti, S.I, Warren, A.L, Nosol, K, Roepe, P.D, Potter, C.S, Carragher, B, Kossiakoff, A.A, Quick, M, Fidock, D.A, Mancia, F. | | Deposit date: | 2019-10-05 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and drug resistance of the Plasmodium falciparum transporter PfCRT.
Nature, 576, 2019
|
|
8E38
 
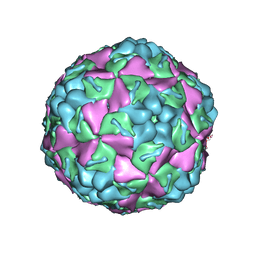 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
6XCC
 
 | |
8T6I
 
 | | Structure of VHH-Fab complex with engineered Crystal Kappa region | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8E2Y
 
 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | Genome polyprotein, VP1, VP2 | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
8E3A
 
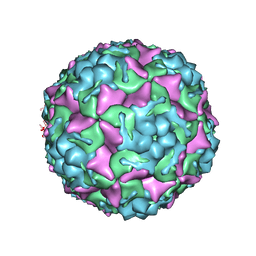 | | Purification of Enterovirus A71, strain 4643, WT capsid | | Descriptor: | VP1, VP2, VP3, ... | | Authors: | Catching, A, Capponi, S, Andino, R. | | Deposit date: | 2022-08-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | A tradeoff between enterovirus A71 particle stability and cell entry.
Nat Commun, 14, 2023
|
|
6MG8
 
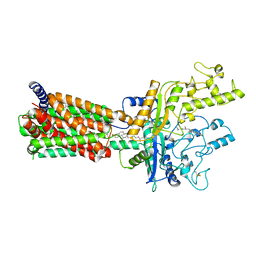 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
6U52
 
 | | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB) Complexed with Sudan ebolavirus Nucleoprotein C-terminal Domain 634-738 | | Descriptor: | Anti-Sudan ebolavirus Nucleoprotein Single Domain Antibody Sudan B (SB), CHLORIDE ION, Nucleoprotein | | Authors: | Taylor, A.B, Sherwood, L.J, Hart, P.J, Hayhurst, A. | | Deposit date: | 2019-08-26 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paratope Duality and Gullying are Among the Atypical Recognition Mechanisms Used by a Trio of Nanobodies to Differentiate Ebolavirus Nucleoproteins.
J.Mol.Biol., 431, 2019
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
