6WHO
 
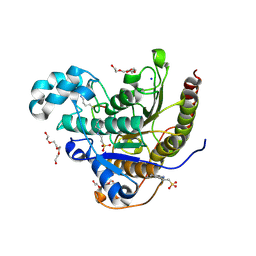 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6ZR0
 
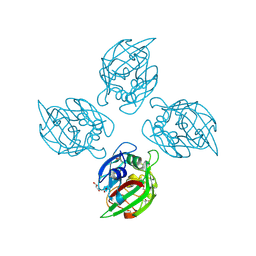 | |
6ZQX
 
 | | Crystal structure of tetrameric fibrinogen-like recognition domain of FIBCD1 with N,N'-diacetyl chitobiose ligand bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CALCIUM ION, ... | | Authors: | Shrive, A.K, Greenhough, T.J, Williams, H.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structures of human immune protein FIBCD1 suggest an extended binding site compatible with recognition of pathogen associated carbohydrate motifs
J.Biol.Chem., 2023
|
|
6WHQ
 
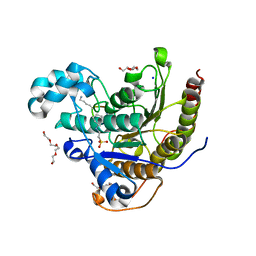 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6ZQY
 
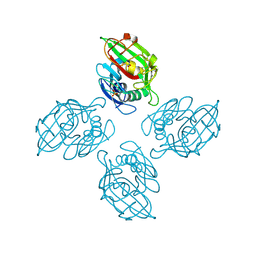 | |
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
8B4S
 
 | | Antimicrobial peptide capitellacin from polychaeta Capitella teleta in DPC (dodecylphosphocholine) micelles, dimeric form | | Descriptor: | BRICHOS domain-containing protein | | Authors: | Mironov, P.A, Reznikova, O.V, Paramonov, A.S, Shenkarev, Z.O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Dimerization of the beta-Hairpin Membrane-Active Cationic Antimicrobial Peptide Capitellacin from Marine Polychaeta: An NMR Structural and Thermodynamic Study.
Biomolecules, 14, 2024
|
|
8E20
 
 | |
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WRW
 
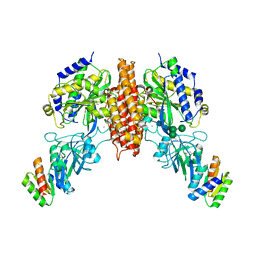 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | Authors: | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZS7
 
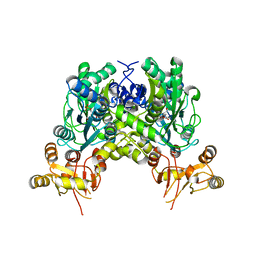 | | Crystal structure of delta466-491 cystathionine beta-synthase from Toxoplasma gondii with L-cysteine | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, Cystathionine beta-synthase | | Authors: | Fernandez-Rodriguez, C, Oyenarte, I, Conter, C, Gonzalez-Recio, I, Quintana, I, Martinez-Chantar, M, Astegno, A, Martinez-Cruz, L.A. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the unique conformation of cystathionine beta-synthase from Toxoplasma gondii .
Comput Struct Biotechnol J, 19, 2021
|
|
6TWV
 
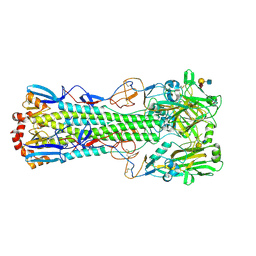 | | Crystal structure of the haemagglutinin mutant (Gln226Leu) from an H10N7 seal influenza virus isolated in Germany with human receptor analogue, 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Hemagglutinin HA1, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-13 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6WKM
 
 | |
6UBC
 
 | | Crystal structure of a GH128 (subgroup VII) oligosaccharide-binding protein from Cryptococcus neoformans (CnGH128_VII) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Santos, C.R, Costa, P.A.C.R, Souza, B.P, Vieira, P.S, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6M3Y
 
 | | Crystal structure of pilus adhesin, SpaC from Lactobacillus rhamnosus GG - open conformation | | Descriptor: | MAGNESIUM ION, Pilus assembly protein | | Authors: | Kant, A, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-29 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of lactobacillar SpaC reveals an atypical five-domain pilus tip adhesin: Exposing its substrate-binding and assembly in SpaCBA pili.
J.Struct.Biol., 211, 2020
|
|
6ZU2
 
 | | CML1 crystal structure in complex with H-type 1 trisaccharide | | Descriptor: | Mucin-binding lectin 1, SULFATE ION, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Varrot, A, Bleuler-Martinez, S. | | Deposit date: | 2020-07-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-function relationship of a novel fucoside-binding fruiting body lectin from Coprinopsis cinerea exhibiting nematotoxic activity.
Glycobiology, 32, 2022
|
|
8DXH
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 5-fluoroquinazolin-4-ol at W266 site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, 5-fluoranyl-3,4-dihydroquinazolin-4-ol, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6ZVC
 
 | |
6ZVB
 
 | |
6UEO
 
 | | Structure of A. thaliana TBP-AC mismatch DNA site | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*CP*CP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TATA-box-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA mismatches reveal conformational penalties in protein-DNA recognition.
Nature, 587, 2020
|
|
8DX8
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-chloro-6-fluorophenethylamine at the 415 site | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-chloro-6-fluorophenyl)ethan-1-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
6OF0
 
 | | Structural basis for multidrug recognition and antimicrobial resistance by MTRR, an efflux pump regulator from Neisseria Gonorrhoeae | | Descriptor: | HTH-type transcriptional regulator MtrR, PHOSPHATE ION | | Authors: | Beggs, G.A, Kumaraswami, M, Shafer, W, Brennan, R.G. | | Deposit date: | 2019-03-28 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Biochemical, and In Vivo Characterization of MtrR-Mediated Resistance to Innate Antimicrobials by the Human Pathogen Neisseria gonorrhoeae .
J.Bacteriol., 201, 2019
|
|
8BV9
 
 | | Acylphosphatase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, GLYCEROL, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First 3-D structural evidence of a native-like intertwined dimer in the acylphosphatase family.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
6M7J
 
 | | Mycobacterium tuberculosis RNAP with RbpA/us fork and Corallopyronin | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2018-08-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding.
Nature, 565, 2019
|
|
8G64
 
 | | Heme-bound flavodoxin FldH from Fusobacterium nucleatum | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Chan, A.C, Wolthers, K.R, Murphy, M.E. | | Deposit date: | 2023-02-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new member of the flavodoxin superfamily from Fusobacterium nucleatum that functions in heme trafficking and reduction of anaerobilin.
J.Biol.Chem., 299, 2023
|
|
