6ZF5
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 1-[3-[(4-butylphenyl)sulfonyl-(2-hydroxy-2-oxoethyl)amino]phenyl]-5-cyclopropyl-pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF1
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl(phenylsulfonyl)amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
8GCC
 
 | | T. cruzi topoisomerase II alpha bound to dsDNA and the covalent inhibitor CT1 | | Descriptor: | 2-{3-[(Z)-iminomethyl]-1H-1,2,4-triazol-1-yl}-1-{(3M)-3-[2-(trifluoromethyl)phenyl]-6H-pyrrolo[3,4-b]pyridin-6-yl}ethan-1-one, DNA (28-MER), DNA topoisomerase 2 | | Authors: | Schenk, A, Deniston, C, Noeske, J. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cyanotriazoles are selective topoisomerase II poisons that rapidly cure trypanosome infections.
Science, 380, 2023
|
|
7OJX
 
 | | E2 UBE2K covalently linked to donor Ub, acceptor di-Ub, and RING E3 primed for K48-linked Ub chain synthesis | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), E3 ubiquitin-protein ligase RNF38, Polyubiquitin-B, ... | | Authors: | Majorek, K.A, Nakasone, M.A, Huang, D.T. | | Deposit date: | 2021-05-17 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of UBE2K-Ub/E3/polyUb reveals mechanisms of K48-linked Ub chain extension.
Nat.Chem.Biol., 18, 2022
|
|
6ZF3
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, SULFATE ION, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF4
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(2,3,5,6-tetramethylphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6ZF7
 
 | | Keap1 kelch domain bound to a small molecule inhibitor of the Keap1-Nrf2 protein-protein interaction | | Descriptor: | 5-cyclopropyl-1-[3-[2-hydroxy-2-oxoethyl-(3-methoxyphenyl)sulfonyl-amino]phenyl]pyrazole-4-carboxylic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1, ... | | Authors: | Narayanan, D, Bach, A, Gajhede, M. | | Deposit date: | 2020-06-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Deconstructing Noncovalent Kelch-like ECH-Associated Protein 1 (Keap1) Inhibitors into Fragments to Reconstruct New Potent Compounds.
J.Med.Chem., 64, 2021
|
|
6YXB
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (space group P21) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6YX6
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant (no morpholine) | | Descriptor: | FLAVIN MONONUCLEOTIDE, Multi-sensor hybrid histidine kinase, SULFATE ION | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
6W2T
 
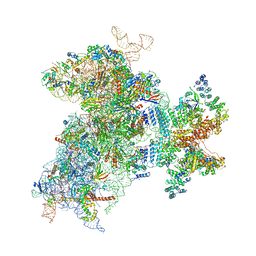 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
8GAJ
 
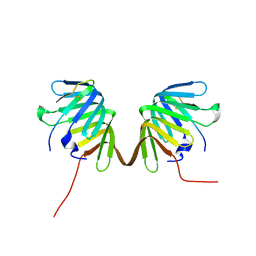 | | Crystal Structure of E. coli LptA in complex with Podisus maculiventris Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Huynh, K, Kibrom, A, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
8GAL
 
 | | Crystal Structure of the E. coli LptA in complex with Murgantia histrionica Thanatin | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Kibrom, A, Huynh, K, Donald, B, Zhou, P. | | Deposit date: | 2023-02-23 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
6YX4
 
 | | Structure of Chloroflexus aggregans flavin based fluorescent protein (CagFbFP) Q148K variant with morpholine | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Multi-sensor hybrid histidine kinase, ... | | Authors: | Remeeva, A, Nazarenko, V, Kovalev, K, Gushchin, I. | | Deposit date: | 2020-04-30 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | The molecular basis of spectral tuning in blue- and red-shifted flavin-binding fluorescent proteins.
J.Biol.Chem., 296, 2021
|
|
7QDD
 
 | |
7O2F
 
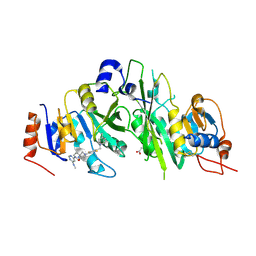 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 22 (UZH2) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-2,5-bis(fluoranyl)phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6PWB
 
 | | Rigid body fitting of flagellin FlaB, and flagellar coiling proteins, FcpA and FcpB, into a 10 Angstrom structure of the asymmetric flagellar filament purified from Leptospira biflexa Patoc WT cells resolved via subtomogram averaging | | Descriptor: | Flagellar coiling protein A (FcpA), Flagellar coiling protein B (FcpB), Flagellin B1 (FlaB1) | | Authors: | Gibson, K.H, Sindelar, C.V, Trajtenberg, F, Buschiazzo, A, San Martin, F, Mechaly, A. | | Deposit date: | 2019-07-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.83 Å) | | Cite: | An asymmetric sheath controls flagellar supercoiling and motility in the leptospira spirochete.
Elife, 9, 2020
|
|
8FLA
 
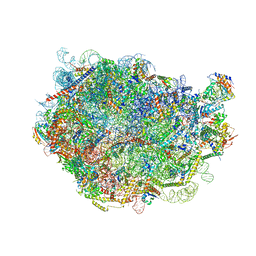 | |
7O2E
 
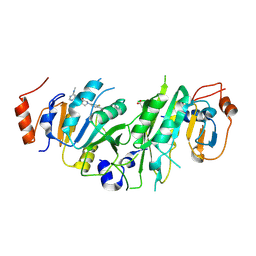 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 21 (ADO_AD_089) | | Descriptor: | 4-[4-[(4,4-dimethylpiperidin-1-yl)methyl]-3-fluoranyl-phenyl]-9-[6-(methylamino)pyrimidin-4-yl]-1,4,9-triazaspiro[5.5]undecan-2-one, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8FL4
 
 | |
7ODA
 
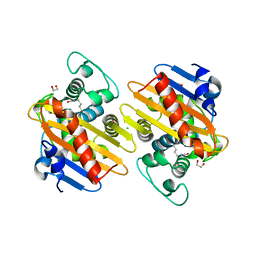 | | OXA-48-like Beta-lactamase OXA-436 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION | | Authors: | Lund, B.A, Thomassen, A.M, Carlsen, T.J.W, Leiros, H.K.S. | | Deposit date: | 2021-04-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Biochemical and biophysical characterization of the OXA-48-like carbapenemase OXA-436.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
8FKT
 
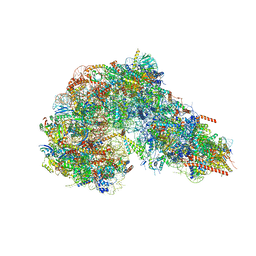 | |
7O09
 
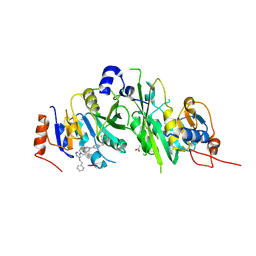 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 7 (ADO_AC_074) | | Descriptor: | 6-[4-[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]-1-oxa-4,9-diazaspiro[5.5]undecan-9-yl]-N-(phenylmethyl)pyrimidin-4-amine, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8FL7
 
 | |
8FLE
 
 | |
7O08
 
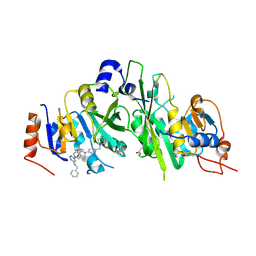 | | Crystal structure of the human METTL3-METTL14 complex bound to Compound 5 (ADO_AB_075) | | Descriptor: | 4-[[[6-[(4,4-dimethylpiperidin-1-yl)methyl]pyridin-3-yl]amino]methyl]-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-4-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Dolbois, A, Caflisch, A. | | Deposit date: | 2021-03-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,9-Triazaspiro[5.5]undecan-2-one Derivatives as Potent and Selective METTL3 Inhibitors.
J.Med.Chem., 64, 2021
|
|
