4UTT
 
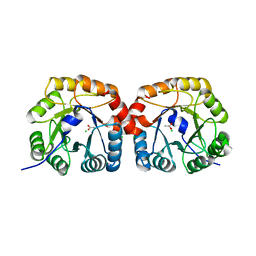 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
4V7S
 
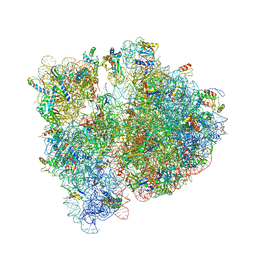 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6U3P
 
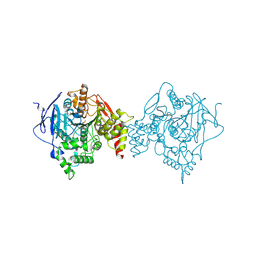 | | Binary complex of native hAChE with oxime reactivator LG-703 | | Descriptor: | (2E,2'E)-N,N'-[1,4-diazepane-1,4-diyldi(ethane-2,1-diyl)]bis[2-(hydroxyimino)acetamide], Acetylcholinesterase, DIMETHYL SULFOXIDE | | Authors: | Kovalevsky, A, Gerlits, O, Radic, Z. | | Deposit date: | 2019-08-22 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Rational design, synthesis, and evaluation of uncharged, "smart" bis-oxime antidotes of organophosphate-inhibited human acetylcholinesterase.
J.Biol.Chem., 295, 2020
|
|
5DVJ
 
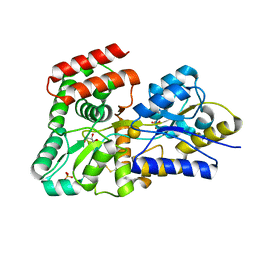 | | Crystal structure of galactose complexed periplasmic glucose binding protein (ppGBP) from P. putida CSV86 | | Descriptor: | Binding protein component of ABC sugar transporter, GLYCEROL, SULFATE ION, ... | | Authors: | Pandey, S, Modak, A, Phale, P.S, Bhaumik, P. | | Deposit date: | 2015-09-21 | | Release date: | 2016-02-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High Resolution Structures of Periplasmic Glucose-binding Protein of Pseudomonas putida CSV86 Reveal Structural Basis of Its Substrate Specificity
J.Biol.Chem., 291, 2016
|
|
6TVQ
 
 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVU
 
 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Transmembrane protein gp41 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
6TVW
 
 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Envelope glycoprotein, Transmembrane protein gp41,Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C, Koksch, B. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
2QYA
 
 | | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri | | Descriptor: | Uncharacterized conserved protein | | Authors: | Bonanno, J.B, Zhang, A, Bain, K.T, Adams, J, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-14 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of an uncharacterized conserved protein from Methanopyrus kandleri.
To be Published
|
|
4V56
 
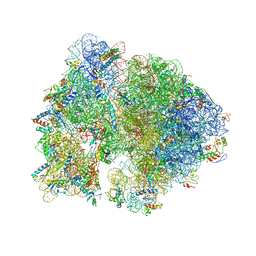 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
6UCH
 
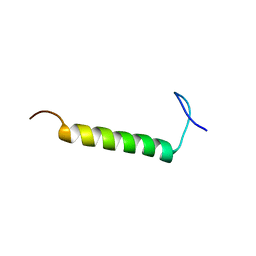 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
4UT4
 
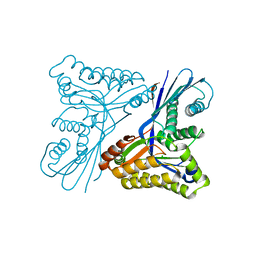 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | Descriptor: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
8UAC
 
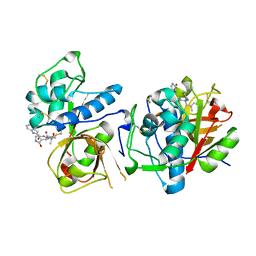 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19.
Med, 5, 2024
|
|
5E68
 
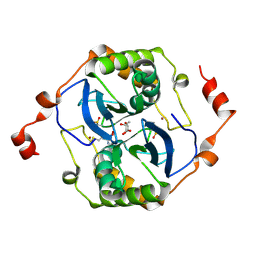 | | High resolution crystal structure of LuxS - Quorum sensor molecular complex from Salmonella typhi at 1.58 Angstroms | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, METHIONINE, S-ribosylhomocysteine lyase, ... | | Authors: | Perumal, P, Raina, R, Arockiasamy, A, SundaraBaalaji, N. | | Deposit date: | 2015-10-09 | | Release date: | 2016-10-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | High resolution crystal structure of LuxS - Quorum sensor molecular complex from Salmonella typhi at 1.58 Angstroms
To Be Published
|
|
4V9N
 
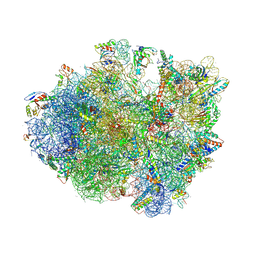 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
6KTL
 
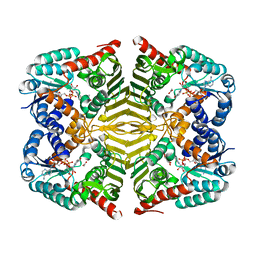 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
2F9H
 
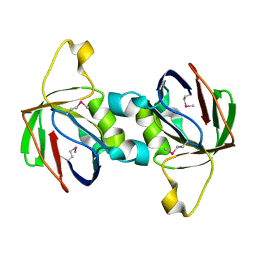 | | The Crystal Structure of PTS System IIA Component from Enterococcus faecalis V583 | | Descriptor: | PTS system, IIA component | | Authors: | Kim, Y, Quartey, P, Moy, S, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal Structure of PTS System IIA Component from Enterococcus faecalis V583
To be Published, 2006
|
|
4UX7
 
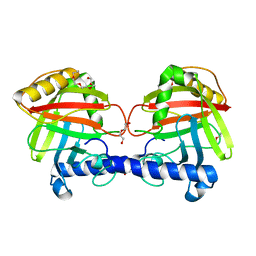 | | Structure of a Clostridium difficile sortase | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, PUTATIVE PEPTIDASE C60B, ... | | Authors: | Chambers, C.J, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2014-08-19 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and Function of a Clostridium Difficile Sortase Enzyme.
Sci.Rep., 5, 2015
|
|
8TNN
 
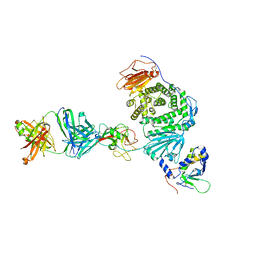 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
2F7R
 
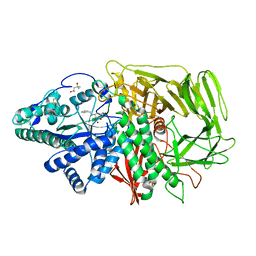 | | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol | | Descriptor: | (1R,2R,3S,4S,5R)-5-(BENZYLAMINO)CYCLOPENTANE-1,2,3,4-TETROL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2005-12-01 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Golgi alpha-mannosidase II complex with benzyl-aminocyclopentitetrol
To be Published
|
|
5EEB
 
 | | Apo form of thermostable aldehyde dehydrogenase from Pyrobaculum sp. 1860 | | Descriptor: | Aldehyde dehydrogenase | | Authors: | Petrova, T.E, Bezsudnova, E.Y, Boyko, K.M, Mardanov, A.V, Gumerov, V.M, Ravin, N.V, Popov, V.O. | | Deposit date: | 2015-10-22 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.038 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
8TRM
 
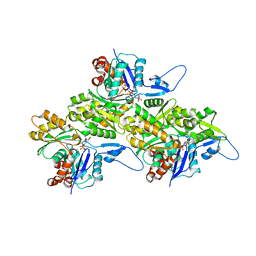 | | Actin 1 from T. gondii in filaments bound to MgADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, MAGNESIUM ION | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
6U0R
 
 | |
8TRN
 
 | | Actin 1 from T. gondii in filaments bound to MgADP and jasplakinolide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, Jasplakinolide, ... | | Authors: | Hvorecny, K.L, Sladewski, T.E, Heaslip, A.T, Kollman, J.M. | | Deposit date: | 2023-08-09 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Toxoplasma gondii actin filaments are tuned for rapid disassembly and turnover.
Nat Commun, 15, 2024
|
|
2QKP
 
 | | Crystal structure of C-terminal domain of SMU_1151c from Streptococcus mutans | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Uncharacterized protein | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Wu, B, Bain, K, Gheyi, T, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of C-terminal domain of SMU_1151c from Streptococcus mutans.
To be Published
|
|
4V8J
 
 | | Crystal structure of the bacterial ribosome ram mutation G347U. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fagan, C.E, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2011-12-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Reorganization of an intersubunit bridge induced by disparate 16S ribosomal ambiguity mutations mimics an EF-Tu-bound state.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
