7T6Q
 
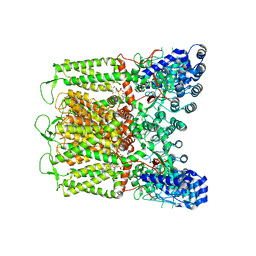 | | Cryo-EM structure of TRPV5 T709D with PI(4,5)P2 in nanodiscs | | Descriptor: | Transient receptor potential cation channel subfamily V member 5, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Fluck, E.C, Yazici, A.T, Rohacs, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2021-12-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of TRPV5 regulation by physiological and pathophysiological modulators.
Cell Rep, 39, 2022
|
|
3EOQ
 
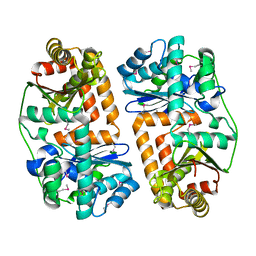 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | Descriptor: | Putative zinc protease | | Authors: | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-09-29 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
5DAD
 
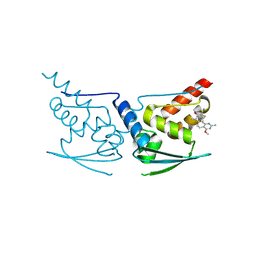 | | Crystal Structure of Human KEAP1 BTB Domain in Complex with Small Molecule TX64014 | | Descriptor: | (6aS,7S,10aS)-8-hydroxy-4-methoxy-2,7,10a-trimethyl-5,6,6a,7,10,10a-hexahydrobenzo[h]quinazoline-9-carbonitrile, Kelch-like ECH-associated protein 1 | | Authors: | Huerta, C, Jiang, X, Trevino, I, Bender, C.F, Swinger, K.K, Stoll, V.S, Ferguson, D.A, Thomas, P.J, Probst, B, Dulubova, I, Visnick, M, Wigley, W.C. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Characterization of novel small-molecule NRF2 activators: Structural and biochemical validation of stereospecific KEAP1 binding.
Biochim.Biophys.Acta, 1860, 2016
|
|
4RHZ
 
 | | Crystal structure of Cry23Aa1 and Cry37Aa1 binary protein complex | | Descriptor: | CALCIUM ION, Cry23AA1, Cry37AA1, ... | | Authors: | Rydel, T.J, Williams, J.M, Brown, G.R, Guzov, V.M, Sturman, E.J, Evdokimov, A. | | Deposit date: | 2014-10-03 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Associated Bacillus thuringiensis Binary Protein Complex of Cry23Aa1 and Cry37Aa1: Crystal Structure, Insecticidal Data, and Pore Formation Modeling.
To be Published
|
|
2IC5
 
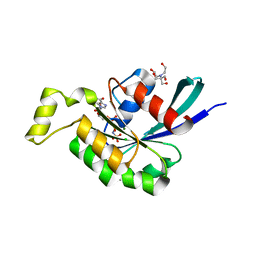 | | Crystal structure of human RAC3 grown in the presence of Gpp(NH)p. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ugochukwu, E, Yang, X, Zao, Y, Elkins, J, Gileadi, C, Burgess, N, Colebrook, S, Gileadi, O, Fedorov, O, Bunkoczi, G, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-09-12 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human RAC3 grown in the presence of Gpp(NH)p.
To be Published
|
|
3EVQ
 
 | |
2IHY
 
 | | Structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter | | Descriptor: | ABC transporter, ATP-binding protein, SULFATE ION | | Authors: | McGrath, T.E, Yu, C.S, Romanov, V, Lam, R, Dharamsi, A, Virag, C, Mansoury, K, Thambipillai, D, Richards, D, Guthrie, J, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2006-09-27 | | Release date: | 2007-09-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Staphylococcus aureus putative ATPase subunit of an ATP-binding cassette (ABC) transporter
To be Published
|
|
2IP4
 
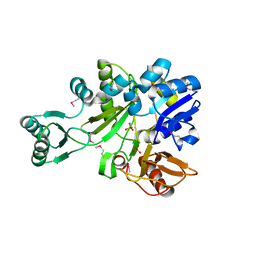 | | Crystal Structure of Glycinamide Ribonucleotide Synthetase from Thermus thermophilus HB8 | | Descriptor: | Phosphoribosylamine--glycine ligase, SULFATE ION | | Authors: | Sampei, G, Baba, S, Kanagawa, M, Yanai, H, Ishii, T, Kawai, H, Fukai, Y, Ebihara, A, Nakagawa, N, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-11 | | Release date: | 2007-10-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria
J.Biochem., 148, 2010
|
|
6Q1F
 
 | | Atomic structure of the Human Herpesvirus 6B Capsid and Capsid-Associated Tegument Complexes | | Descriptor: | Large structural phosphoprotein, Major capsid protein, Small capsomere-interacting protein, ... | | Authors: | Zhang, Y.B, Liu, W, Li, Z.H, Kumar, V, Alvarez-Cabrera, A.L, Leibovitch, E, Cui, Y.X, Mei, Y, Bi, G.Q, Jacobson, S, Zhou, Z.H. | | Deposit date: | 2019-08-03 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes.
Nat Commun, 10, 2019
|
|
4RR2
 
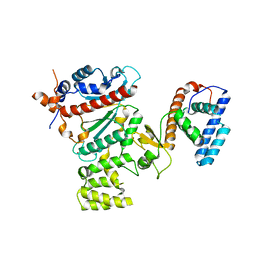 | | Crystal structure of human primase | | Descriptor: | DNA primase large subunit, DNA primase small subunit, IRON/SULFUR CLUSTER, ... | | Authors: | Baranovskiy, A.G, Gu, J, Suwa, Y, Babayeva, N.D, Tahirov, T.H. | | Deposit date: | 2014-11-05 | | Release date: | 2015-01-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the human primase.
J.Biol.Chem., 290, 2015
|
|
4RXU
 
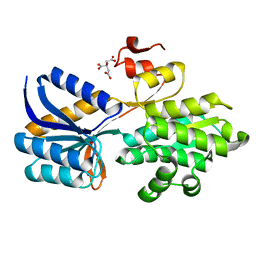 | | Crystal structure of carbohydrate transporter solute binding protein CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158, in complex with D-glucose | | Descriptor: | CITRIC ACID, Periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of sugar transporter CAUR_1924 from Chloroflexus aurantiacus, Target EFI-511158
To be Published
|
|
7RVB
 
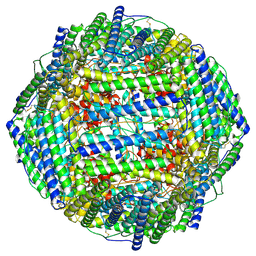 | | High resolution map of molecular chaperone Artemin | | Descriptor: | Ferritin | | Authors: | Parvate, A.D, Powell, S.M, Brookreason, J.T, Novikova, I.V, Evans, J.E. | | Deposit date: | 2021-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Cryo-EM structure of the diapause chaperone artemin.
Front Mol Biosci, 9, 2022
|
|
2I00
 
 | | Crystal structure of acetyltransferase (GNAT family) from Enterococcus faecalis | | Descriptor: | Acetyltransferase, GNAT family | | Authors: | Zhang, R, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-08-09 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the acetyltransferase (GNAT family) from Enterococcus faecalis
To be Published, 2006
|
|
6I5Z
 
 | | Papaver somniferum O-methyltransferase | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE | | Authors: | Davies, G.J, Cabry, M.P, Offen, W.A. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
2I5T
 
 | | Crystal Structure of hypothetical protein LOC79017 from Homo sapiens | | Descriptor: | Protein C7orf24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-12 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of Homo sapiens protein LOC79017.
Proteins, 70, 2008
|
|
1SXH
 
 | | apo structure of B. megaterium transcription regulator | | Descriptor: | Glucose-resistance amylase regulator | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
6I81
 
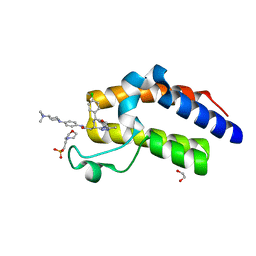 | | Crystal Structure of the second bromodomain of BRD2 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2, ... | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal Structure of the second bromodomain of BRD2 in complex with RT56
To Be Published
|
|
1TFW
 
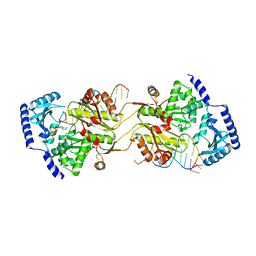 | |
7T9J
 
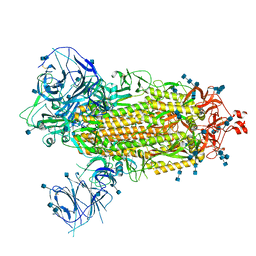 | | Cryo-EM structure of the SARS-CoV-2 Omicron spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
7T9K
 
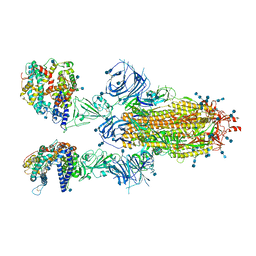 | | Cryo-EM structure of SARS-CoV-2 Omicron spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2021-12-19 | | Release date: | 2021-12-29 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex.
Science, 375, 2022
|
|
1SXI
 
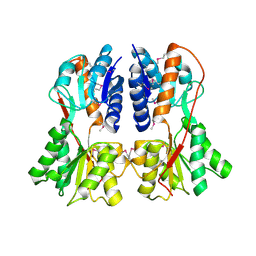 | | Structure of apo transcription regulator B. megaterium | | Descriptor: | Glucose-resistance amylase regulator, MAGNESIUM ION | | Authors: | Schumacher, M.A, Allen, G.S, Diel, M, Seidel, G, Hillen, W, Brennan, R.G. | | Deposit date: | 2004-03-30 | | Release date: | 2004-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on the apo transcription factor form B. megaterium
Cell(Cambridge,Mass.), 118, 2004
|
|
7TAV
 
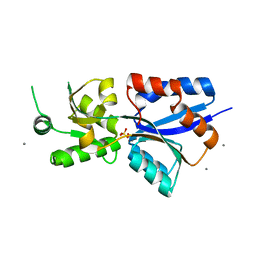 | | Crystal Structure of the PBP2_YvgL_like protein Lmo1041 from Listeria monocytogene | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Grimshaw, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-21 | | Release date: | 2021-12-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the PBP2_YvgL_like protein Lmo1041 from Listeria monocytogenes
To Be Published
|
|
6I7Y
 
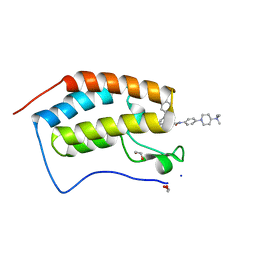 | | Crystal Structure of the first bromodomain of BRD4 in complex with RT56 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, [1-[4-[2-[(4~{S})-6-(4-chlorophenyl)-8-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]ethanoylamino]phenyl]piperidin-4-yl]-trimethyl-azanium | | Authors: | Picaud, S, Traquete, R, Bernardes, G.J.L, Newman, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Filippakopoulos, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-19 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal Structure of the first bromodomain of BRD4 in complex with RT56
To Be Published
|
|
7AJZ
 
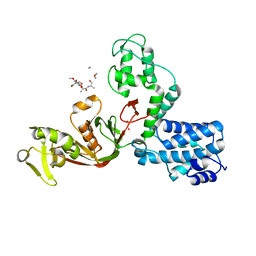 | | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide) | | Descriptor: | 1,2-ETHANEDIOL, L,D-transpeptidase YcbB, NAG-NAM(tetrapeptide), ... | | Authors: | Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The X-ray Structure of L,D-transpeptidase LdtA from Vibrio cholerae in complex with NAG-NAM(tetrapeptide)
To Be Published
|
|
7AJ9
 
 | |
