5FSJ
 
 | | Structure of thermolysin prepared by the 'soak-and-freeze' method under 45 bar of oxygen pressure | | Descriptor: | CALCIUM ION, LYSINE, OXYGEN MOLECULE, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, P, Carpentier, P. | | Deposit date: | 2016-01-06 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
6F9P
 
 | | Crystal structure of the Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Re(II) | | Descriptor: | CHLORIDE ION, GLYCEROL, L-lysine 4-hydroxylase, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-12-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
7MYH
 
 | | Ubiquitin variant UbV.k.2 in complex with Ube2k | | Descriptor: | GLYCEROL, Ubiquitin variant UbV.k.2, Ubiquitin-conjugating enzyme E2 K | | Authors: | Middleton, A.J, Day, C.L, Teyra, J, Sidhu, S.S. | | Deposit date: | 2021-05-21 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Identification of Ubiquitin Variants That Inhibit the E2 Ubiquitin Conjugating Enzyme, Ube2k.
Acs Chem.Biol., 16, 2021
|
|
6VGT
 
 | |
4YO9
 
 | | HKU4 3CLpro unbound structure | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3C-like proteinase, ACETATE ION, ... | | Authors: | St John, S.E, Mesecar, A. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting zoonotic viruses: Structure-based inhibition of the 3C-like protease from bat coronavirus HKU4-The likely reservoir host to the human coronavirus that causes Middle East Respiratory Syndrome (MERS).
Bioorg.Med.Chem., 23, 2015
|
|
8UUK
 
 | | Pendrin in apo | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Hoang, A, Zhou, M. | | Deposit date: | 2023-11-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of anion exchange and small-molecule inhibition of pendrin.
Nat Commun, 15, 2024
|
|
6EXA
 
 | |
8SQW
 
 | | particulate methane monooxygenase crosslinked with 2,2,2-trifluoroethanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Ammonia monooxygenase/methane monooxygenase, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8SR1
 
 | | particulate methane monooxygenase crosslinked with 4,4,4-trifluorobutanol bound | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dihexanoyl-sn-glycero-3-phosphocholine, 4,4,4-trifluorobutan-1-ol, ... | | Authors: | Tucci, F.J, Rosenzweig, A.C. | | Deposit date: | 2023-05-05 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Product analog binding identifies the copper active site of particulate methane monooxygenase.
Nat Catal, 6, 2023
|
|
8GJ3
 
 | | E. coli clamp loader on primed template DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase III subunit delta, DNA polymerase III subunit delta', ... | | Authors: | Oakley, A.J, Xu, Z.-Q, Dixon, N.E. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural characterisation of the complete cycle of sliding clamp loading in E. coli
To Be Published
|
|
2WIH
 
 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-848125 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
6QWP
 
 | | Crystal structure of T7 bacteriophage portal protein, 13mer, closed valve | | Descriptor: | Portal protein | | Authors: | Fabrega-Ferrer, M, Cuervo, A, Machon, C, Fernandez, F.J, Perez-Luque, R, Pous, J, Vega, M.C, Carrascosa, J.L, Coll, M. | | Deposit date: | 2019-03-06 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Nat Commun, 10, 2019
|
|
8GJ1
 
 | |
6B37
 
 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol | | Descriptor: | 1,3-benzothiazol-2-ol, DIMETHYL SULFOXIDE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-21 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 1,3-benzothiazol-2-ol
To Be Published
|
|
4YQY
 
 | | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028) (TARGET EFI-513936) in its APO form | | Descriptor: | MAGNESIUM ION, Putative Dehydrogenase | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Koss, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2015-03-13 | | Release date: | 2015-03-25 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Crystal Structure of a putative Dehydrogenase from Sulfitobacter sp. (COG1028, TARGET EFI-513936) in its APO form
To be published
|
|
5JFD
 
 | | Thrombin in complex with (S)-N-(2-(aminomethyl)-5-chlorobenzyl)-1-((benzylsulfonyl)-D-arginyl)pyrrolidine-2-carboxamide | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G. | | Deposit date: | 2016-04-19 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Strategies for Late-Stage Optimization: Profiling Thermodynamics by Preorganization and Salt Bridge Shielding.
J.Med.Chem., 62, 2019
|
|
6B3L
 
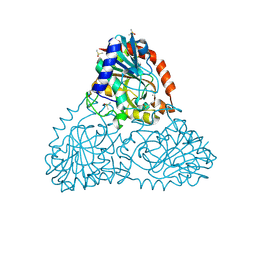 | | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2,3-dihydro-1H-inden-2-amine | | Descriptor: | DIMETHYL SULFOXIDE, INDAN-2-AMINE, Purine nucleoside phosphorylase | | Authors: | Faheem, M, Neto, J.B, Collins, P, Pearce, N.M, Valadares, N.F, Bird, L, Pereira, H.M, Delft, F.V, Barbosa, J.A.R.G. | | Deposit date: | 2017-09-22 | | Release date: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Purine Nucleoside Phosphorylase Isoform 2 from Schistosoma mansoni in complex with 2,3-dihydro-1H-inden-2-amine
To Be Published
|
|
8GJ2
 
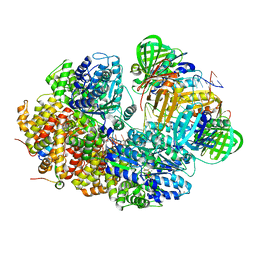 | |
8GJ0
 
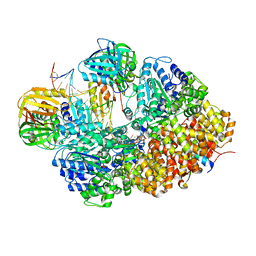 | |
5FT0
 
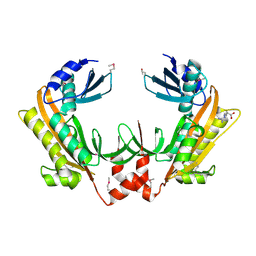 | | Crystal structure of gp37(Dip) from bacteriophage phiKZ | | Descriptor: | ARGININE, GP37, POTASSIUM ION | | Authors: | Van den Bossche, A, Hardwick, S.W, Ceyssens, P.J, Hendrix, H, Voet, M, Dendooven, T, Bandyra, K.J, De Maeyer, M, Aertsen, A, Noben, J.P, Luisi, B.F, Lavigne, R. | | Deposit date: | 2016-01-08 | | Release date: | 2016-08-03 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural elucidation of a novel mechanism for the bacteriophage-based inhibition of the RNA degradosome.
Elife, 5, 2016
|
|
6ETK
 
 | |
6B5V
 
 | | Structure of TRPV5 in complex with econazole | | Descriptor: | 1-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, CALCIUM ION, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Hughes, T.E.T, Lodowski, D.T, Huynh, K.W, Yazici, A, del Rosario, J, Kapoor, A, Basak, S, Samanta, A, Chakrapani, S, Zhou, Z.H, Filizola, M, Rohacs, T, Han, S, Moiseenkova-Bell, V.Y. | | Deposit date: | 2017-09-29 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5JGU
 
 | | Spin-Labeled T4 Lysozyme Construct R119V1 | | Descriptor: | CHLORIDE ION, Endolysin, PHOSPHATE ION, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-04-20 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.468 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
7X55
 
 | |
6F6Z
 
 | | Mouse Thymidylate Synthase Cocrystallized with N(4)OHdCMP and Soaked in Methylenetetrahydrofolate | | Descriptor: | (2~{S})-2-[[4-[[(6~{R})-2-azanyl-4-oxidanylidene-5,6,7,8-tetrahydro-1~{H}-pteridin-6-yl]methyl-methyl-amino]phenyl]carbonylamino]pentanedioic acid, 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), Thymidylate synthase | | Authors: | Wilk, P, Jarmula, A, Maj, P, Rode, W. | | Deposit date: | 2017-12-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Mechanism of Thymidylate Synthase Inhibition by N4-Hydroxy-dCMP in View of Spectrophotometric and Crystallographic Studies
Int J Mol Sci, 22, 2021
|
|
