5K52
 
 | |
4DSL
 
 | | Crystal structure of fragment DNA polymerase I from Bacillus stearothermophilus with duplex DNA and Calcium | | Descriptor: | DNA (5'-D(*G*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*C)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3'), DNA polymerase, ... | | Authors: | Gan, J.H, Abdur, R, Liu, H.H, Sheng, J, Caton-Willians, J, Soares, A.S, Huang, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Biochemical and structural insights into the fidelity of bacillus stearothermophilus DNA polymerase
To be Published
|
|
5CQA
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with N-methyl-2,3-dihydrothieno[3,4-b][1,4]dioxine-5-carboxamide (SGC - Diamond I04-1 fragment screening)
To be published
|
|
7A8P
 
 | | Structure of human mitochondrial RNA polymerase in complex with IMT inhibitor. | | Descriptor: | (3~{R})-1-[(2~{R})-2-[4-(2-chloranyl-4-fluoranyl-phenyl)-2-oxidanylidene-chromen-7-yl]oxypropanoyl]piperidine-3-carboxylic acid, DNA-directed RNA polymerase, mitochondrial | | Authors: | Hillen, H.S, Bonekamp, N, Peter, B, Felser, A, Bergbrede, T, Choidas, A, Horn, M, Unger, A, di Lucrezia, R, Atanassov, I, Li, X, Koch, U, Menninger, S, Boros, J, Habenberger, P, Giavalisco, P, Cramer, P, Denzel, M, Nussbaumer, P, Klebl, B, Falkenberg, M, Gustafsson, C.M, Larsson, N.G. | | Deposit date: | 2020-08-30 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Small-molecule inhibitors of human mitochondrial DNA transcription.
Nature, 588, 2020
|
|
7G1G
 
 | | Crystal Structure of human FABP4 in complex with 2-(2-hydroxy-3,5,5,8,8-pentamethyl-3,4,4a,6,7,8a-hexahydro-1H-naphthalen-2-yl)acetic acid | | Descriptor: | FORMIC ACID, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Hochstetler-A, D, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8E3T
 
 | |
7FWQ
 
 | | Crystal Structure of human FABP4 binding site mutated to that of FABP3 in complex with 6-chloro-4-(2-chlorophenoxy)-2-methylquinoline-3-carboxylic acid | | Descriptor: | 6-chloro-4-(2-chlorophenoxy)-2-methylquinoline-3-carboxylic acid, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Conte, A, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a human FABP4 binding site mutated to that of FABP3 complex
To be published
|
|
7AB1
 
 | | Crystal structure of MerTK kinase domain in complex with Gilteritinib | | Descriptor: | 6-ethyl-3-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-5-(oxan-4-ylamino)pyrazine-2-carboxamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Overman, R.C, Rawlins, P.B, Truman, C, Underwood, E, Warwicker, J, Winter-Holt, J. | | Deposit date: | 2020-09-05 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A-loop interactions in Mer tyrosine kinase give rise to inhibitors with two-step mechanism and long residence time of binding.
Biochem.J., 477, 2020
|
|
8DQH
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 24 hours of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
6UVZ
 
 | | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis | | Descriptor: | Amidohydrolase 2, CITRIC ACID, NONAETHYLENE GLYCOL | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-11-04 | | Release date: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Amidohydrolase 2 from Bifidobacterium longum subsp. infantis
To Be Published
|
|
6S96
 
 | |
5MDN
 
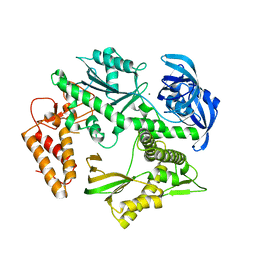 | | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis | | Descriptor: | DNA polymerase, MAGNESIUM ION | | Authors: | Guo, J, Zhang, W, Coker, A.R, Wood, S.P, Cooper, J.B, Rashid, N, Akhtar, M. | | Deposit date: | 2016-11-12 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the family B DNA polymerase from the hyperthermophilic archaeon Pyrobaculum calidifontis.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8DQI
 
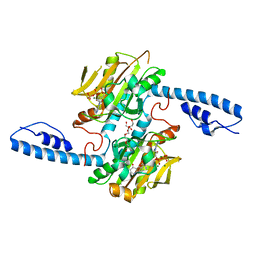 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to ATP and acridone after 2- weeks of crystal growth | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
4DD5
 
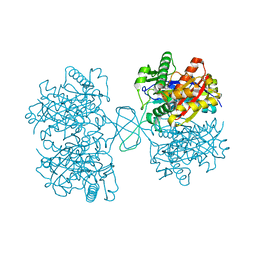 | | Biosynthetic Thiolase (ThlA1) from Clostridium difficile | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-18 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biosynthetic Thiolase (ThlA1) from Clostridium difficile
To be Published
|
|
8AK4
 
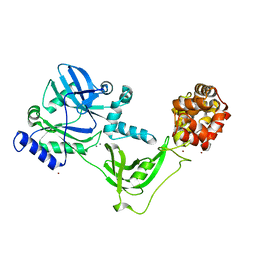 | | Structure of the C-terminally truncated NAD+-dependent DNA ligase from the poly-extremophile Deinococcus radiodurans | | Descriptor: | DNA ligase, MANGANESE (II) ION, ZINC ION | | Authors: | Fernandes, A, Williamson, A.K, Matias, P.M, Moe, E. | | Deposit date: | 2022-07-29 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structure/function studies of the NAD + -dependent DNA ligase from the poly-extremophile Deinococcus radiodurans reveal importance of the BRCT domain for DNA binding.
Extremophiles, 27, 2023
|
|
7RKV
 
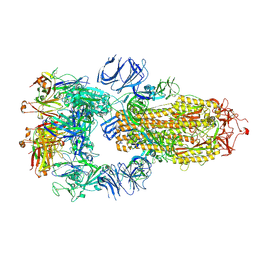 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
4Z37
 
 | |
6S8K
 
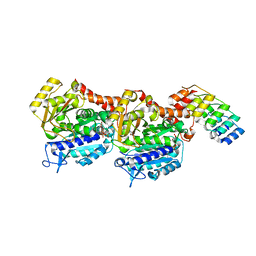 | | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to two Tubulin Isotypes | | Descriptor: | (3Z,6Z)-3-benzylidene-6-[(5-tert-butyl-1H-imidazol-4-yl)methylidene]piperazine-2,5-dione, Designed ankyrin repeat protein (DARPIN) D1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sharma, A, Olieric, N, Steinmetz, M. | | Deposit date: | 2019-07-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure, Thermodynamics, and Kinetics of Plinabulin Binding to Two Tubulin Isotypes
Chem, 5, 2019
|
|
8ANV
 
 | |
4YYH
 
 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6XRX
 
 | | Crystal structure of the mosquito protein AZ1 as an MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Foo, A.C.Y. | | Deposit date: | 2020-07-14 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mosquito protein AEG12 displays both cytolytic and antiviral properties via a common lipid transfer mechanism.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4DG5
 
 | |
4YYN
 
 | | Crystal structure of TAF1 BD2 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
8QK9
 
 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8E5W
 
 | | Crystal structure of dehydroalanine Hip1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PALMITIC ACID, Protease, ... | | Authors: | Goldfarb, N.E, Brooks, C.L, Ostrov, D.A. | | Deposit date: | 2022-08-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
