6I9Q
 
 | | Structure of the mouse CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, CHLORIDE ION | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural differences between the ectodomains of murine and human CD98hc.
Proteins, 87, 2019
|
|
6AOS
 
 | |
7M9G
 
 | | HIV-1 Protease (I84V) in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-18
To Be Published
|
|
6S5R
 
 | | Cfucosylated second generation peptide dendrimer SBD6 bound to Fucose binding Lectin LecB (PA-IIL) from Pseudomonas aeruginosa at 2.08 Angstrom resolution, incomplete structure | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fucose-binding lectin, ... | | Authors: | Baeriswyl, S, Stocker, A, Reymond, J.-L. | | Deposit date: | 2019-07-02 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | X-Ray Crystal Structure of a Second Generation Peptide Dendrimer in Complex with Pseudomonas aeruginosa Lectin LecB
Helv.Chim.Acta, 2019
|
|
7M9H
 
 | | HIV-1 Protease (I84V) in Complex with LR2-20 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with LR2-20
To Be Published
|
|
7UFP
 
 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | Mur ligase middle domain protein, SULFATE ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2022-03-23 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
7UGS
 
 | |
4RE4
 
 | | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Beta-mannosidase/beta-glucosidase, ... | | Authors: | Tankrathok, A, Iglesias-Fernandez, J, Williams, R.J, Hakki, Z, Robinson, R.C, Hrmova, M, Rovira, C, Williams, S.J, Ketudat Cairns, J.R. | | Deposit date: | 2014-09-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Single Glycosidase Harnesses Different Pyranoside Ring Transition State Conformations for Hydrolysis of Mannosides and Glucosides
ACS CATALYSIS, 5, 2015
|
|
7M9J
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-68 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
8A8Y
 
 | |
6AR1
 
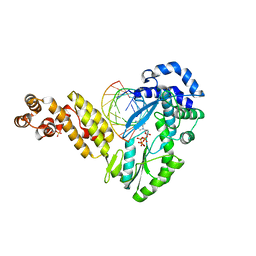 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
4REE
 
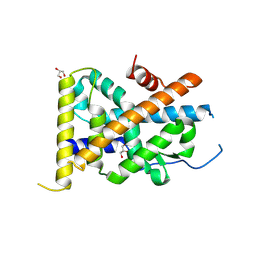 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
7RAE
 
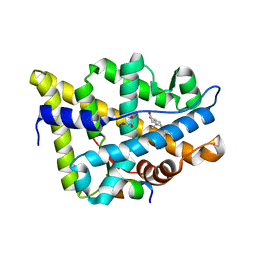 | | AncAR1 - progesterone - Tif2 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Ancestral androgen receptor, GLYCEROL, ... | | Authors: | Ortlund, E.A. | | Deposit date: | 2021-07-01 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | AncAR1 - DHT - Tif2
To Be Published
|
|
6ARJ
 
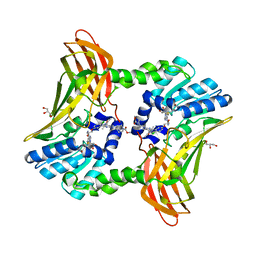 | | Crystal structure of CARM1 with EPZ022302 and SAH | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
5XMX
 
 | | Co-crystal structure of Inhibitor compound in complex with human PPARdelta LBD | | Descriptor: | (E)-6-[2-[[[4-(furan-2-yl)phenyl]carbonyl-methyl-amino]methyl]phenoxy]-4-methyl-hex-4-enoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Lakshminarasimhan, A, Rani, S.T, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel highly selective peroxisome proliferator-activated receptor delta (PPAR delta) modulators with pharmacokinetic properties suitable for once-daily oral dosing.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6VE7
 
 | | The inner junction complex of Chlamydomonas reinhardtii doublet microtubule | | Descriptor: | Cilia- and flagella-associated protein 20, FAP276, FAP52, ... | | Authors: | Khalifa, A.A.Z, Ichikawa, M, Bui, K.H. | | Deposit date: | 2019-12-30 | | Release date: | 2020-02-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The inner junction complex of the cilia is an interaction hub that involves tubulin post-translational modifications.
Elife, 9, 2020
|
|
6E6E
 
 | | DGY-06-116, a novel and selective covalent inhibitor of SRC kinase | | Descriptor: | N-(2-chloro-6-methylphenyl)-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-4-{[2-(propanoylamino)phenyl]amino}pyrimidine-5-carboxamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Gurbani, D, Bera, A, Westover, K. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Characterization of a Covalent Inhibitor of Src Kinase.
Front Mol Biosci, 7, 2020
|
|
4RC1
 
 | | Structure of the methanofuran/methanopterin biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii with PRPP | | Descriptor: | PHOSPHATE ION, UPF0264 protein MJ1099 | | Authors: | Bobik, T.A, Morales, E.J, Cascio, D, Sawaya, M.R, Yeates, T.O, Rasche, M.E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the methanofuran/methanopterin-biosynthetic enzyme MJ1099 from Methanocaldococcus jannaschii.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
7MMT
 
 | |
7JLG
 
 | |
7MMV
 
 | |
7JL8
 
 | |
7MMS
 
 | |
7UIY
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6EBL
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
