4QVT
 
 | | Crystal structure of predicted N-acyltransferase (ypeA) in complex with acetyl-CoA from Escherichia coli | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase YpeA, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Winsor, G, Dubrovska, I, Shuvalova, L, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Crystal structure of predicted N-acyltransferase (ypeA) in complex with acetyl-CoA from Escherichia coli
To be Published
|
|
5G1M
 
 | | Crystal structure of NagZ from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, BETA-HEXOSAMINIDASE, CHLORIDE ION, ... | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-03-28 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
4XRA
 
 | | Salmonella typhimurium AhpC T43S mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Perkins, A, Brereton, A.E, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
8PU4
 
 | | FaRel2 bound to the ATP analogue, APCPP | | Descriptor: | MAGNESIUM ION, RelA/SpoT domain-containing protein, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[[oxidanyl(phosphonooxy)phosphoryl]methyl]phosphinic acid | | Authors: | Garcia-Pino, A, Talavera Perez, A, Dominguez Molina, L. | | Deposit date: | 2023-07-16 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | FaRel2 bound to the ATP analogue, APCPP
To Be Published
|
|
6A4K
 
 | | Human antibody 32D6 Fab in complex with H1N1 influenza A virus HA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Lee, C.C, Ko, T.P, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2018-06-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An Effective Neutralizing Antibody Against Influenza Virus H1N1 from Human B Cells.
Sci Rep, 9, 2019
|
|
6RVF
 
 | | Crystal structure of hCA II in complex with Urea, N-(1,3-dihydro-1-hydroxy-2,1-benzoxaborol-6-yl)-N'-phenyl | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1$l^{4}-benzoxaborol-6-yl]-3-phenyl-urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Exploring benzoxaborole derivatives as carbonic anhydrase inhibitors: a structural and computational analysis reveals their conformational variability as a tool to increase enzyme selectivity.
J Enzyme Inhib Med Chem, 34, 2019
|
|
7M5H
 
 | | Crystal structure of conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase | | Authors: | Nocek, B, Wu, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-03-23 | | Release date: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of conserved protein from Enterococcus faecalis V583
To Be Published
|
|
5G3F
 
 | | Preserving Metallic Sites Affected by Radiation Damage the CuT2 CAse in Thermus Thermophilus Multicopper Oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Ruiz-Arellano, R, Diaz, A, Rosas, E, Rudino, E. | | Deposit date: | 2016-04-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Preserving Metallic Sites Affected by Radiation Damage the Cut2 Case in Thermus Thermophilus Multicopper Oxidase
To be Published
|
|
4XRD
 
 | | Salmonella typhimurium AhpC W169F mutant | | Descriptor: | Alkyl hydroperoxide reductase subunit C, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Perkins, A, Nelson, K, Parsonage, D, Poole, L, Karplus, P.A. | | Deposit date: | 2015-01-21 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Experimentally Dissecting the Origins of Peroxiredoxin Catalysis.
Antioxid.Redox Signal., 28, 2018
|
|
4QYI
 
 | | 1.95 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' with HEPES molecule in the active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Hypoxanthine phosphoribosyltransferase, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-24 | | Release date: | 2014-08-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom resolution crystal structure of a hypoxanthine-guanine phosphoribosyltransferase (hpt-2) from Bacillus anthracis str. 'Ames Ancestor' with HEPES molecule in the active site
To be Published
|
|
7MBK
 
 | |
8FMD
 
 | |
4XW5
 
 | | X-ray structure of PKAc with ATP, CP20, calcium ions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
8DJ3
 
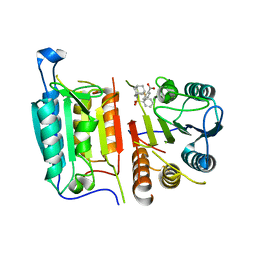 | | Caspase-7 bound to novel allosteric inhibitor | | Descriptor: | 2-[(2-{[(3s,5s,7s)-adamantan-1-yl]sulfamoyl}phenyl)sulfanyl]benzoic acid, Caspase-7 | | Authors: | Propp, J, Kalenkiewicz, A, Kathryn, F.H, Spies, M.A. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Tuning of Caspase-7: Establishing the Nexus of Structure and Catalytic Power.
Chemistry, 29, 2023
|
|
6E53
 
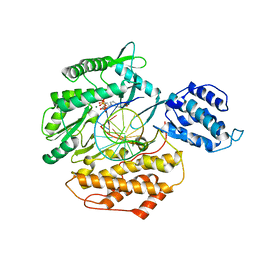 | | Structure of TERT in complex with a novel telomerase inhibitor | | Descriptor: | MAGNESIUM ION, RNA/DNA hairpin, Telomerase reverse transcriptase, ... | | Authors: | Hernandez-Sanchez, W, Huang, W, Plucinsky, B, Garcia-Vazquez, N, Berdis, A.J, Skordalakes, E, Taylor, D.J. | | Deposit date: | 2018-07-19 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A non-natural nucleotide uses a specific pocket to selectively inhibit telomerase activity.
Plos Biol., 17, 2019
|
|
6E5E
 
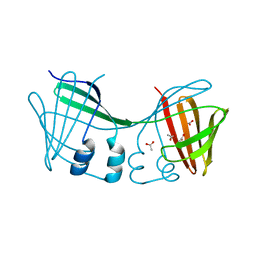 | |
7QLB
 
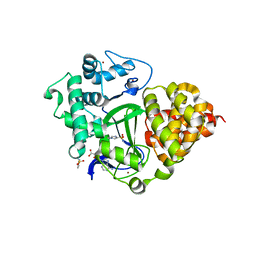 | | SMYD3 in complex with fragment FL06268 | | Descriptor: | 1-methylimidazole-4-sulfonamide, Histone-lysine N-methyltransferase SMYD3, S-ADENOSYLMETHIONINE, ... | | Authors: | Lund, B.A, Cederfelt, D, Dobritzsch, D. | | Deposit date: | 2021-12-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of fragments targeting SMYD3 using highly sensitive kinetic and multiplexed biosensor-based screening
Rsc Med Chem, 2024
|
|
6E5Q
 
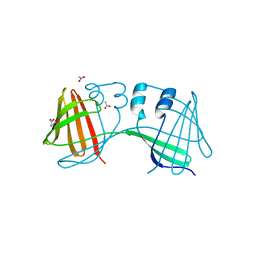 | |
6A4J
 
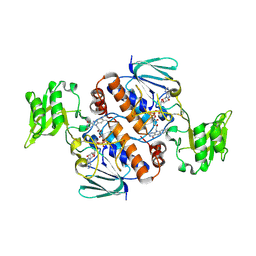 | | Crystal structure of Thioredoxin reductase 2 from Staphylococcus aureus | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Bose, M, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
5C0G
 
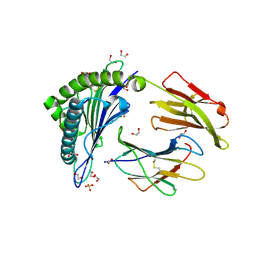 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
6S41
 
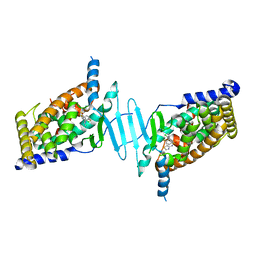 | | CRYSTAL STRUCTURE OF PXR IN COMPLEX WITH XPC-7455 | | Descriptor: | 4-[[(1~{S})-1-[2,5-bis(fluoranyl)phenyl]ethyl]amino]-5-chloranyl-2-fluoranyl-~{N}-(1,3-thiazol-4-yl)benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Focken, T, Maskos, K, Griessner, A, Krapp, S. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of CNS-Penetrant Aryl Sulfonamides as Isoform-Selective NaV1.6 Inhibitors with Efficacy in Mouse Models of Epilepsy.
J.Med.Chem., 62, 2019
|
|
6E5Y
 
 | | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Argininosuccinate synthase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Resolution Crystal Structure of Argininosuccinate Synthase from Bordetella pertussis in Complex with AMP.
To Be Published
|
|
6S4H
 
 | | The crystal structure of glycogen phosphorylase in complex with 8 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
4XXF
 
 | | L-fuculose 1-phosphate aldolase from Glaciozyma antarctica PI12 | | Descriptor: | Fuculose-1-phosphate aldolase, ZINC ION | | Authors: | Jaafar, N.R, Abu Bakar, F.D, Abdul Murad, A.M, Illias, R, Littler, D, Beddoe, T, Rossjohn, J, Mahadi, M.N. | | Deposit date: | 2015-01-30 | | Release date: | 2015-02-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of fuculose aldolase from the Antarctic psychrophilic yeast Glaciozyma antarctica PI12.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6S52
 
 | | The crystal structure of glycogen phosphorylase in complex with 14 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(5-phenyl-1,2,3,4-tetrazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
