5KCI
 
 | | Crystal Structure of HTC1 | | 分子名称: | GLYCEROL, SULFATE ION, Uncharacterized protein YPL067C, ... | | 著者 | Martin, R.M, Horowitz, S, Koepnick, B, Cooper, S, Flatten, J, Rogawski, D.S, Koropatkin, N.M, Beinlich, F.R.M, Players, F, Students, U.M, Popovic, Z, Baker, D, Khatib, F, Bardwell, J.C.A. | | 登録日 | 2016-06-06 | | 公開日 | 2016-09-21 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.833 Å) | | 主引用文献 | Determining crystal structures through crowdsourcing and coursework.
Nat Commun, 7, 2016
|
|
8ABP
 
 | |
5NFX
 
 | | Deinococcus radiodurans BphP PAS-GAF Y263F mutant | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, ACETATE ION, ... | | 著者 | Takala, H, Westenhoff, S, Ihalainen, J.A. | | 登録日 | 2017-03-16 | | 公開日 | 2018-04-18 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.34 Å) | | 主引用文献 | On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome.
J. Biol. Chem., 293, 2018
|
|
7U5G
 
 | | ACS122 Fab | | 分子名称: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | 著者 | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | 登録日 | 2022-03-02 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
6WQV
 
 | | GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose | | 分子名称: | 1,2-ETHANEDIOL, Endoglucanase, NITRATE ION, ... | | 著者 | Bianchetti, C.M, Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | 登録日 | 2020-04-29 | | 公開日 | 2020-11-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
6TP4
 
 | | Crystal structure of the Orexin-1 receptor in complex with ACT-462206 | | 分子名称: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, (2~{S})-~{N}-(3,5-dimethylphenyl)-1-(4-methoxyphenyl)sulfonyl-pyrrolidine-2-carboxamide, Orexin receptor type 1, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-12 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3.011 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6FQV
 
 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | 著者 | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6TPG
 
 | | Crystal structure of the Orexin-2 receptor in complex with EMPA at 2.74 A resolution | | 分子名称: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.741 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6FTB
 
 | | Staphylococcus aureus monofunctional glycosyltransferase in complex with moenomycin | | 分子名称: | (2R)-2,3-dihydroxypropyl dodecanoate, 1,2-ETHANEDIOL, MOENOMYCIN, ... | | 著者 | Punekar, A.S, Dowson, C.J, Roper, D.I. | | 登録日 | 2018-02-20 | | 公開日 | 2018-06-27 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The role of the jaw subdomain of peptidoglycan glycosyltransferases for lipid II polymerization.
Cell Surf, 2, 2018
|
|
4Z13
 
 | | Recombinantly expressed latent aurone synthase (polyphenol oxidase) co-crystallized with hexatungstotellurate(VI) and soaked in H2O2 | | 分子名称: | 6-tungstotellurate(VI), Aurone synthase, COPPER (II) ION, ... | | 著者 | Molitor, C, Mauracher, S.G, Rompel, A. | | 登録日 | 2015-03-26 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Aurone synthase is a catechol oxidase with hydroxylase activity and provides insights into the mechanism of plant polyphenol oxidases.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5NJ5
 
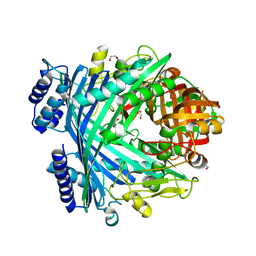 | | E. coli Microcin-processing metalloprotease TldD/E with phosphate bound | | 分子名称: | 1,2-ETHANEDIOL, Metalloprotease PmbA, Metalloprotease TldD, ... | | 著者 | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | 登録日 | 2017-03-28 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
6FX1
 
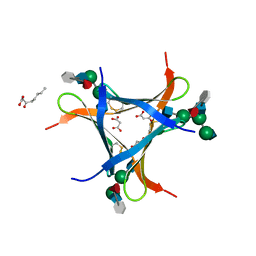 | | Crystal structure of Pholiota squarrosa lectin in complex with an octasaccharide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, ... | | 著者 | Cabanettes, A, Varrot, A. | | 登録日 | 2018-03-08 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6YWM
 
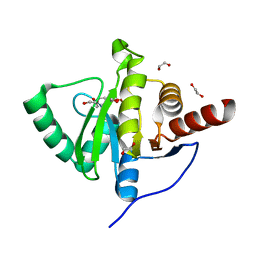 | | Crystal structure of SARS-CoV-2 (Covid-19) NSP3 macrodomain in complex with MES | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | 著者 | Ni, X, Schroeder, M, Olieric, V, Sharpe, E.M, Wojdyla, J.A, Wang, M, Knapp, S, Chaikuad, A, Structural Genomics Consortium (SGC) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural Insights into Plasticity and Discovery of Remdesivir Metabolite GS-441524 Binding in SARS-CoV-2 Macrodomain.
Acs Med.Chem.Lett., 12, 2021
|
|
6Z47
 
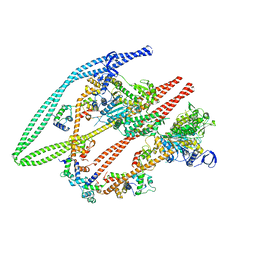 | | Smooth muscle myosin shutdown state heads region | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin heavy chain 11, ... | | 著者 | Scarff, C.A, Carrington, G, Casas Mao, D, Chalovich, J.M, Knight, P.J, Ranson, N.A, Peckham, M. | | 登録日 | 2020-05-22 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Structure of the shutdown state of myosin-2.
Nature, 588, 2020
|
|
6X4D
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(cyclopropylmethyl)-7-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-8-methyl-2-naphthonitrile (JLJ678), a Non-nucleoside Inhibitor | | 分子名称: | 5-(cyclopropylmethyl)-7-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-8-methylnaphthalene-2-carbonitrile, Reverse transcriptase/ribonuclease H, SULFATE ION, ... | | 著者 | Chan, A.H, Duong, V.N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2020-05-22 | | 公開日 | 2020-07-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural investigation of 2-naphthyl phenyl ether inhibitors bound to WT and Y181C reverse transcriptase highlights key features of the NNRTI binding site.
Protein Sci., 29, 2020
|
|
7U9V
 
 | | Integrin alaphIIBbeta3 complex with BMS4-1 | | 分子名称: | (4-{[(5S)-3-(4-carbamimidoylphenyl)-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 10E5 Fab heavy chain, 10E5 light chain, ... | | 著者 | Zhu, J, Lin, F.-Y, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-11 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25492167 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
5NPI
 
 | |
8VK2
 
 | | X-ray crystal structure of human IgE 4C8 Fab | | 分子名称: | IgE 4C8 heavy chain, IgE 4C8 light chain | | 著者 | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | 登録日 | 2024-01-08 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.508 Å) | | 主引用文献 | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 2024
|
|
5KLF
 
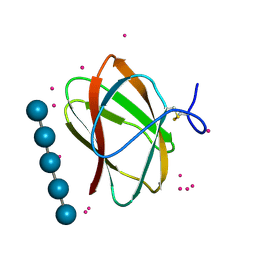 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose and gadolinium ion | | 分子名称: | Carbohydrate binding module E1, GADOLINIUM ATOM, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | 登録日 | 2016-06-24 | | 公開日 | 2016-09-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
6X1B
 
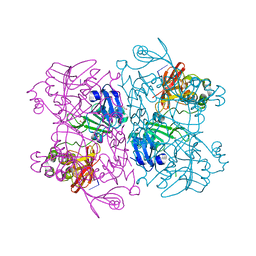 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with the Product Nucleotide GpU. | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-R(*GP*U)-3'), PHOSPHATE ION, ... | | 著者 | Kim, Y, Maltseva, N, Jedrzejczak, R, Welk, L, Endres, M, Chang, C, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-18 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8PTI
 
 | | CRYSTAL STRUCTURE OF A Y35G MUTANT OF BOVINE PANCREATIC TRYPSIN INHIBITOR | | 分子名称: | BOVINE PANCREATIC TRYPSIN INHIBITOR | | 著者 | Housset, D, Kim, K.-S, Fuchs, J, Woodward, C, Wlodawer, A. | | 登録日 | 1990-12-17 | | 公開日 | 1991-04-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a Y35G mutant of bovine pancreatic trypsin inhibitor.
J.Mol.Biol., 220, 1991
|
|
6FY6
 
 | | Concerted dynamics of metallo-base pairs in an A/B-form helical transition (major species) | | 分子名称: | DNA (5'-D(*CP*GP*TP*CP*TP*CP*AP*TP*GP*AP*TP*AP*CP*G)-3')_major, MERCURY (II) ION | | 著者 | Schmidt, O.P, Jurt, S, Johannsen, S, Karimi, A, Sigel, R.K.O, Luedtke, N.W. | | 登録日 | 2018-03-11 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Concerted dynamics of metallo-base pairs in an A/B-form helical transition.
Nat Commun, 10, 2019
|
|
7U9F
 
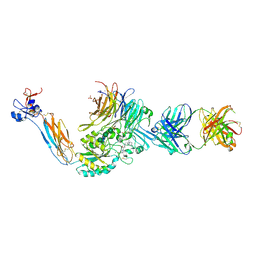 | | Integrin alaphIIBbeta3 complex with BMS compound 4 in Mn2+ | | 分子名称: | (4-{[(5S)-3-{4-[(E)-imino(4-methylpiperazin-1-yl)methyl]phenyl}-4,5-dihydro-1,2-oxazol-5-yl]methyl}piperazin-1-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Lin, F.-Y, Zhu, J, Zhu, J, Springer, T.A. | | 登録日 | 2022-03-10 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.70000529 Å) | | 主引用文献 | A general chemical principle for creating closure-stabilizing integrin inhibitors.
Cell, 185, 2022
|
|
5KLC
 
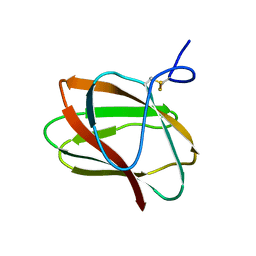 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome | | 分子名称: | Carbohydrate binding module E1 | | 著者 | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | 登録日 | 2016-06-24 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.746 Å) | | 主引用文献 | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
7TJC
 
 | |
