6MHR
 
 | | Structure of the human 4-1BB / Urelumab Fab complex | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, MALONATE ION, ... | | 著者 | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | 登録日 | 2018-09-18 | | 公開日 | 2018-11-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6MI2
 
 | | Structure of the human 4-1BB / Utomilumab Fab complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | 著者 | Kimberlin, C.R, Chin, S.M, Roe-Zurz, Z, Xu, A, Yang, Y. | | 登録日 | 2018-09-19 | | 公開日 | 2018-11-21 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structure of the 4-1BB/4-1BBL complex and distinct binding and functional properties of utomilumab and urelumab.
Nat Commun, 9, 2018
|
|
6FKF
 
 | | Chloroplast F1Fo conformation 1 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | 著者 | Hahn, A, Vonck, J, Mills, D.J, Meier, T, Kuehlbrandt, W. | | 登録日 | 2018-01-24 | | 公開日 | 2018-05-23 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure, mechanism, and regulation of the chloroplast ATP synthase.
Science, 360, 2018
|
|
6BNN
 
 | | Crystal structure of V278E-glyoxalase I mutant from Zea mays in space group P4(1)2(1)2 | | 分子名称: | COBALT (II) ION, FORMIC ACID, GLUTATHIONE, ... | | 著者 | Alvarez, C.E, Agostini, R.B, Gonzalez, J.M, Drincovich, M.F, Campos Bermudez, V.A, Klinke, S. | | 登録日 | 2017-11-17 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Deciphering the number and location of active sites in the monomeric glyoxalase I of Zea mays.
Febs J., 286, 2019
|
|
6FK4
 
 | | Structure of 3' phosphatase NExo (WT) from Neisseria bound to DNA substrate | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*TP*AP*GP*CP*GP*AP*AP*GP*CP*TP*AP*GP*A)-3'), Exodeoxyribonuclease III | | 著者 | Silhan, J, Zhao, Q, Boura, E, Thomson, H, Foster, A, Tang, C.M, Freemont, P.S, Baldwin, G.S. | | 登録日 | 2018-01-23 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.297 Å) | | 主引用文献 | Structural basis for recognition and repair of the 3'-phosphate by NExo, a base excision DNA repair nuclease from Neisseria meningitidis.
Nucleic Acids Res., 46, 2018
|
|
6MIA
 
 | | Crystal structure of CTX-M-14 with compound 6 | | 分子名称: | 3-(1H-tetrazol-5-ylmethyl)-5,6,7,8-tetrahydro[1]benzothieno[2,3-d]pyrimidin-4(3H)-one, Beta-lactamase | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2018-09-19 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | 分子名称: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | 著者 | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | 登録日 | 2017-03-23 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
8P9K
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB503 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-4-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)benzamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
7QEV
 
 | | human Connexin 26 at 55mm Hg PCO2, pH7.4:two masked subunits, class D | | 分子名称: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | 著者 | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | 登録日 | 2021-12-03 | | 公開日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
6MJT
 
 | | Azurin 122F/124W/126Re | | 分子名称: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), Azurin, COPPER (II) ION | | 著者 | Takematsu, K, Zalis, S, Gray, H.B, Vlcek, A, Winkler, J.R, Williamson, H, Kaiser, J.T, Heyda, J, Hollas, D. | | 登録日 | 2018-09-21 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Two Tryptophans Are Better Than One in Accelerating Electron Flow through a Protein.
ACS Cent Sci, 5, 2019
|
|
3JVA
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 | | 分子名称: | Dipeptide Epimerase, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Sakai, A, Imker, H, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-09-16 | | 公開日 | 2010-08-11 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6C34
 
 | | Mycobacterium smegmatis DNA flap endonuclease mutant D125N | | 分子名称: | 5'-3' exonuclease, MANGANESE (II) ION | | 著者 | Shuman, S, Goldgur, Y, Carl, A, Uson, M.L. | | 登録日 | 2018-01-09 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Nucleic Acids Res., 46, 2018
|
|
8P9J
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB500 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
4RN6
 
 | | Structure of prethrombin-2 mutant s195a bound to the active site inhibitor argatroban | | 分子名称: | (2R,4R)-4-methyl-1-(N~2~-{[(3S)-3-methyl-1,2,3,4-tetrahydroquinolin-8-yl]sulfonyl}-L-arginyl)piperidine-2-carboxylic acid, Thrombin heavy chain | | 著者 | Pozzi, N, Chen, Z, Zapata, F, Niu, W, Barranco-Medina, S, Pelc, L.A, Di Cera, E. | | 登録日 | 2014-10-23 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
6MJP
 
 | | LptB(E163Q)FGC from Vibrio cholerae | | 分子名称: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-cyclohexylhexyl beta-D-glucopyranoside, ABC transporter ATP-binding protein, ... | | 著者 | Owens, T.W, Kahne, D, Kruse, A.C. | | 登録日 | 2018-09-21 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
5UK8
 
 | | The co-structure of (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine and a rationally designed PI3K-alpha mutant that mimics ATR | | 分子名称: | (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Knapp, M.S, Mamo, M, Elling, R.A. | | 登録日 | 2017-01-20 | | 公開日 | 2017-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
7AG0
 
 | | Complex between the bone morphogenetic protein 2 and its antagonist Noggin | | 分子名称: | Bone morphogenetic protein 2, GLYCEROL, Noggin | | 著者 | Robert, C, Bruck, F, Herman, R, Vandevenne, M, Filee, P, Kerff, F, Matagne, A. | | 登録日 | 2020-09-21 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3.104 Å) | | 主引用文献 | Structural analysis of the interaction between human cytokine BMP-2 and the antagonist Noggin reveals molecular details of cell chondrogenesis inhibition.
J.Biol.Chem., 299, 2023
|
|
8P9F
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB161 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-(2-azanyl-5-methyl-4-oxidanyl-phenyl)hydrazinyl]benzamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
6SLD
 
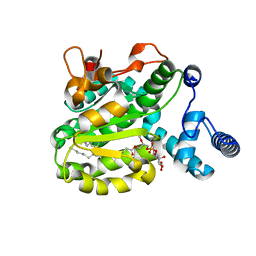 | | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol | | 分子名称: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CRAL-TRIO domain-containing protein YKL091C | | 著者 | Berger, J, Fitz, M, Johnen, P, Shanmugaratnam, S, Hocker, B, Stiel, A.C, Schaaf, G. | | 登録日 | 2019-08-19 | | 公開日 | 2020-08-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol
To Be Published
|
|
5HB8
 
 | |
8P9L
 
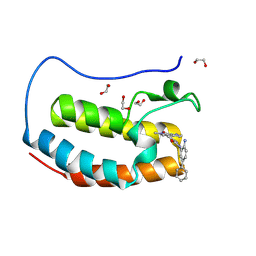 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual BET/HDAC inhibitor NB512 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-azanyl-5-phenyl-phenyl)-5-(6-methyl-7-oxidanylidene-1~{H}-pyrrolo[2,3-c]pyridin-4-yl)pyridine-2-carboxamide | | 著者 | Balourdas, D.I, Bauer, N, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | 登録日 | 2023-06-06 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | Development of Potent Dual BET/HDAC Inhibitors via Pharmacophore Merging and Structure-Guided Optimization.
Acs Chem.Biol., 19, 2024
|
|
6GU3
 
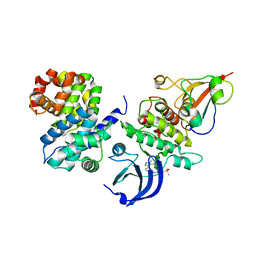 | | CDK1/CyclinB/Cks2 in complex with AZD5438 | | 分子名称: | 4-(2-methyl-3-propan-2-yl-imidazol-4-yl)-~{N}-(4-methylsulfonylphenyl)pyrimidin-2-amine, Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, ... | | 著者 | Wood, D.J, Korolchuk, S, Tatum, N.J, Wang, L.Z, Endicott, J.A, Noble, M.E.M, Martin, M.P. | | 登録日 | 2018-06-19 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Differences in the Conformational Energy Landscape of CDK1 and CDK2 Suggest a Mechanism for Achieving Selective CDK Inhibition.
Cell Chem Biol, 26, 2019
|
|
7QWZ
 
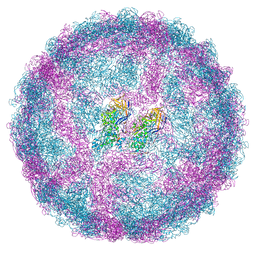 | | Full capsid of Saccharomyces cerevisiae virus L-BCLa | | 分子名称: | Major capsid protein | | 著者 | Grybchuk, D, Prochazkova, M, Fuzik, T, Konovalovas, A, Serva, S, Yurchenko, V, Plevka, P. | | 登録日 | 2022-01-26 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of L-BC virus and its open particle provide insight into Totivirus capsid assembly.
Commun Biol, 5, 2022
|
|
5V3G
 
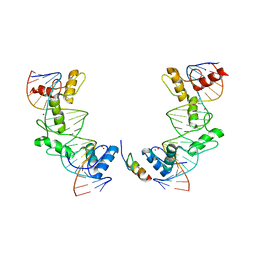 | | PRDM9-allele-C ZnF8-13 | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | 著者 | Patel, A, Cheng, X. | | 登録日 | 2017-03-07 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.416 Å) | | 主引用文献 | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
6GX6
 
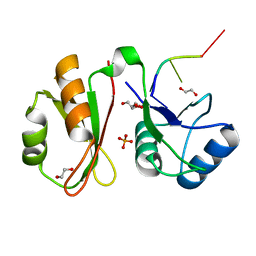 | | Crystal structure of IMP3 RRM12 in complex with RNA (ACAC) | | 分子名称: | 1,2-ETHANEDIOL, Insulin-like growth factor 2 mRNA-binding protein 3, PHOSPHATE ION, ... | | 著者 | Jia, M, Gut, H, Chao, A.J. | | 登録日 | 2018-06-26 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of IMP3 RRM12 recognition of RNA.
RNA, 24, 2018
|
|
