6B4U
 
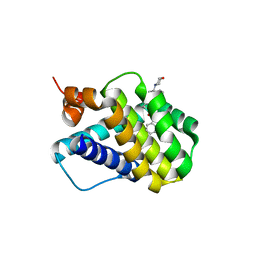 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | 分子名称: | 7-(2-methylphenyl)-1-[2-(morpholin-4-yl)ethyl]-3-{3-[(naphthalen-1-yl)oxy]propyl}-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | 著者 | Judge, R.A, Souers, A.J. | | 登録日 | 2017-09-27 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
5XVK
 
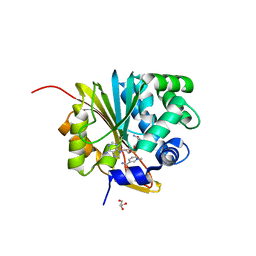 | | Crystal structure of mouse Nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl Nicotinamide (MNA) | | 分子名称: | 3-carbamoyl-1-methylpyridin-1-ium, GLYCEROL, Nicotinamide N-methyltransferase, ... | | 著者 | Swaminathan, S, Birudukota, S, Thakur, M.K, Parveen, R, Kandan, S, Kannt, A, Gosu, R. | | 登録日 | 2017-06-28 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structures of monkey and mouse nicotinamide N-methyltransferase (NNMT) bound with end product, 1-methyl nicotinamide
Biochem. Biophys. Res. Commun., 491, 2017
|
|
6B4H
 
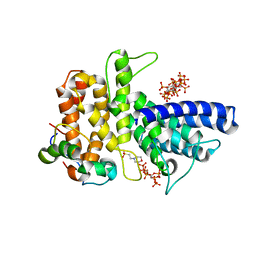 | | Crystal structure of Chaetomium thermophilum Gle1 CTD-Nup42 GBM-IP6 complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INOSITOL HEXAKISPHOSPHATE, Nucleoporin AMO1, ... | | 著者 | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | 登録日 | 2017-09-26 | | 公開日 | 2018-06-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
3LPR
 
 | |
8EWW
 
 | |
5XW3
 
 | |
7JNO
 
 | |
6FAV
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 4.2f-I with 14-3-3sigma | | 分子名称: | (2~{R})-2-[(~{R})-(3-methoxyphenyl)-phenyl-methyl]pyrrolidine, 14-3-3 protein sigma, ACE-ARG-THR-PRO-SEP-LEU-PRO-GLY, ... | | 著者 | Andrei, S.A, Meijer, F.A, Ottmann, C, Milroy, L.G. | | 登録日 | 2017-12-18 | | 公開日 | 2018-05-16 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Inhibition of 14-3-3/Tau by Hybrid Small-Molecule Peptides Operating via Two Different Binding Modes.
ACS Chem Neurosci, 9, 2018
|
|
4TL7
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4Y45
 
 | |
5XWE
 
 | | Structure of a three finger toxin from Ophiophagus hannah venom | | 分子名称: | CHLORIDE ION, GLYCEROL, Weak toxin DE-1 homolog 1, ... | | 著者 | Jobichen, C, Roy, A, Kini, R.M, Sivaraman, J. | | 登録日 | 2017-06-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of a three finger toxin from Ophiophagus hannah venom
To Be Published
|
|
5I00
 
 | | tRNA guanine transglycosylase (TGT) in co-crystallized complex with 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | 分子名称: | 1,2-ETHANEDIOL, 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, CHLORIDE ION, ... | | 著者 | Ehrmann, F.R, Heine, A, Klebe, G. | | 登録日 | 2016-02-03 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Co-crystallization, Isothermal titration calorimetry and nanoESI-MS reveal dimer disturbing inhibitors
To be Published
|
|
6B6H
 
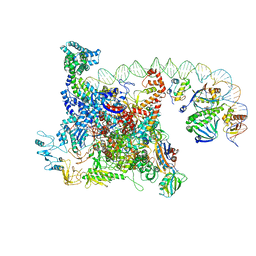 | | The cryo-EM structure of a bacterial class I transcription activation complex | | 分子名称: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | 登録日 | 2017-10-02 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
8W01
 
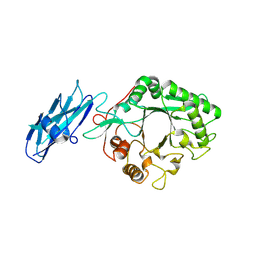 | |
7JLW
 
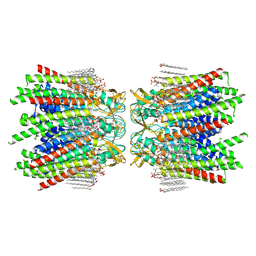 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 1 | | 分子名称: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | 著者 | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | 登録日 | 2020-07-30 | | 公開日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
7JOA
 
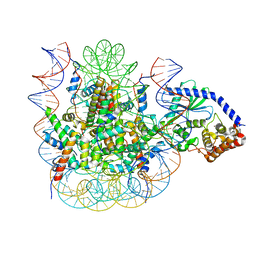 | | 2:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | 著者 | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | 登録日 | 2020-08-06 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
8V8L
 
 | | Switchgrass Chalcone Isomerase | | 分子名称: | Chalcone-flavonone isomerase family protein, GLYCEROL | | 著者 | Lewis, J.A, Kang, C. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
4YEF
 
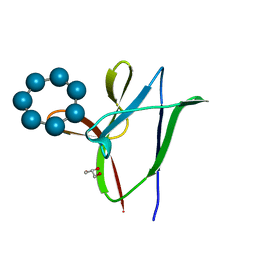 | | beta1 carbohydrate binding module (CBM) of AMP-activated protein kinase (AMPK) in complex with glucosyl-beta-cyclododextrin | | 分子名称: | 5'-AMP-activated protein kinase subunit beta-1, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | 著者 | Mobbs, J, Gorman, M.A, Parker, M.W, Gooley, P.R, Griffin, M. | | 登録日 | 2015-02-24 | | 公開日 | 2015-06-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Determinants of oligosaccharide specificity of the carbohydrate-binding modules of AMP-activated protein kinase.
Biochem.J., 468, 2015
|
|
8V8P
 
 | | Sorghum Chalcone Isomerase | | 分子名称: | 7-HYDROXY-2-(4-HYDROXY-PHENYL)-CHROMAN-4-ONE, Chalcone-flavonone isomerase family protein | | 著者 | Lewis, J.A, Kang, C. | | 登録日 | 2023-12-05 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Structural and Interactional Analysis of the Flavonoid Pathway Proteins: Chalcone Synthase, Chalcone Isomerase and Chalcone Isomerase-like Protein.
Int J Mol Sci, 25, 2024
|
|
4YF6
 
 | | Crystal structure of oxidised Rv1284 | | 分子名称: | Beta-carbonic anhydrase 1, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Hofmann, A. | | 登録日 | 2015-02-25 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.002 Å) | | 主引用文献 | Chemical probing suggests redox-regulation of the carbonic anhydrase activity of mycobacterial Rv1284.
Febs J., 282, 2015
|
|
7UYX
 
 | | Structure of bacteriophage PA1c gp2 | | 分子名称: | Bacteriophage PA1C gp2 | | 著者 | Enustun, E, Deep, A, Gu, Y, Nguyen, K, Chaikeeratisak, V, Armbruster, E, Ghassemian, M, Pogliano, J, Corbett, K.D. | | 登録日 | 2022-05-07 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Identification of the bacteriophage nucleus protein interaction network.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6B7Y
 
 | | Cryo-EM structure of human insulin degrading enzyme | | 分子名称: | Insulin-degrading enzyme | | 著者 | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | 登録日 | 2017-10-05 | | 公開日 | 2017-11-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
7JVK
 
 | |
4YH6
 
 | | Crystal structure of IL1RAPL1 ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yamagata, A, Fukai, S. | | 登録日 | 2015-02-27 | | 公開日 | 2015-05-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Mechanisms of splicing-dependent trans-synaptic adhesion by PTP delta-IL1RAPL1/IL-1RAcP for synaptic differentiation.
Nat Commun, 6, 2015
|
|
8UZB
 
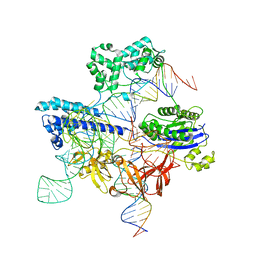 | | Cryo-EM structure of iGeoCas9 in complex with sgRNA and target DNA | | 分子名称: | CRISPR-associated endonuclease Cas9, Non-target strand DNA, RNA (107-MER), ... | | 著者 | Eggers, A.R, Soczek, K.M, Tuck, O.T, Doudna, J.A. | | 登録日 | 2023-11-14 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | Rapid DNA unwinding accelerates genome editing by engineered CRISPR-Cas9.
Cell, 187, 2024
|
|
