6V0Q
 
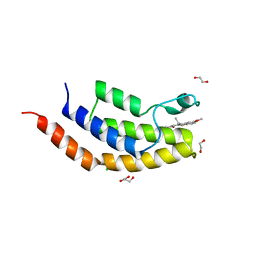 | | Crystal structure of the bromodomain of human BRD7 bound to TG003 | | 分子名称: | (1~{Z})-1-(3-ethyl-5-methoxy-1,3-benzothiazol-2-ylidene)propan-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 7, ... | | 著者 | Karim, M.R, Chan, A, Schonbrunn, E. | | 登録日 | 2019-11-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1H
 
 | | Crystal structure of the bromodomain of human BRD7 bound to bromosporine | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 7, Bromosporine | | 著者 | Chan, A, Karim, M.R, Schonbrunn, E. | | 登録日 | 2019-11-20 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
6V1L
 
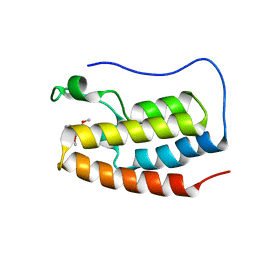 | | Crystal structure of the first bromodomain (BD1) of human BRD4 bound to BI9564 | | 分子名称: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 4 | | 著者 | Chan, A, Karim, M.R, Schonbrunn, E. | | 登録日 | 2019-11-20 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis of Inhibitor Selectivity in the BRD7/9 Subfamily of Bromodomains.
J.Med.Chem., 63, 2020
|
|
1L1F
 
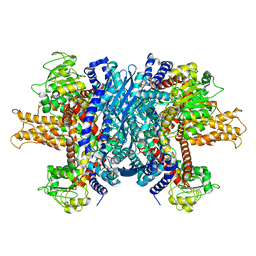 | | Structure of human glutamate dehydrogenase-apo form | | 分子名称: | Glutamate Dehydrogenase 1 | | 著者 | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | 登録日 | 2002-02-15 | | 公開日 | 2002-03-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
4DUI
 
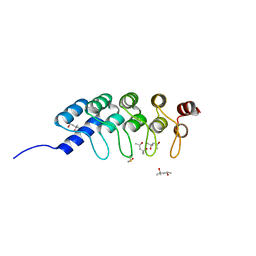 | | DARPIN D1 binding to tubulin beta chain (not in complex) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN) D1 | | 著者 | Pecqueur, L, Duellberg, C, Dreier, B, Wang, Q, Jiang, C, Pluckthun, A, Surrey, T, Gigant, B, Knossow, M. | | 登録日 | 2012-02-22 | | 公開日 | 2013-02-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | An Anti-Tubulin Darpin Caps the Microtubule Plus-End
To be Published
|
|
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | 著者 | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | 登録日 | 2016-05-23 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structural studies of human muscle FBPase
To Be Published
|
|
8TH3
 
 | |
6V3Q
 
 | | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form | | 分子名称: | ISOPROPYL ALCOHOL, Metallo-beta-lactamase FIM-1, ZINC ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-11-26 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of the Metallo-beta-Lactamase FIM-1 from Pseudomonas aeruginosa in the Mono-Zinc Form
To Be Published
|
|
5U4Z
 
 | | Crystal structure of citrus MAF1 in space group P 31 2 1 | | 分子名称: | Repressor of RNA polymerase III transcription, SULFATE ION | | 著者 | Soprano, A.S, Giuseppe, P.O, Nascimento, A.F.Z, Benedetti, C.E, Murakami, M.T. | | 登録日 | 2016-12-06 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal structure of citrus MAF1 in space group P 31 2 1
To Be Published
|
|
5MY0
 
 | | KS-MAT DI-DOMAIN OF MOUSE FAS WITH MALONYL-COA | | 分子名称: | COENZYME A, Fatty acid synthase, MALONYL-COENZYME A | | 著者 | Paithankar, K.S, Rittner, A, Huu, K.V, Grininger, M. | | 登録日 | 2017-01-25 | | 公開日 | 2018-01-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Characterization of the Polyspecific Transferase of Murine Type I Fatty Acid Synthase (FAS) and Implications for Polyketide Synthase (PKS) Engineering.
ACS Chem. Biol., 13, 2018
|
|
6IAS
 
 | |
4R2F
 
 | | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus A6, target EFI-510633, with bound laminaribiose | | 分子名称: | Extracellular solute-binding protein family 1, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-08-11 | | 公開日 | 2014-08-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of sugar transporter ACHL_0255 from Arthrobacter chlorophenolicus, target EFI-510633
To be Published
|
|
1KZT
 
 | |
6SQL
 
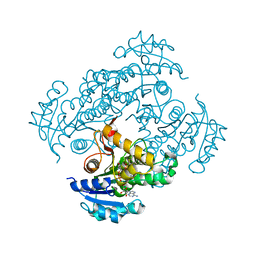 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and N-(3-(aminomethyl)phenyl)-5-chloro-3-methylbenzo[b]thiophene-2-sulfonamide | | 分子名称: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-[3-(aminomethyl)phenyl]-5-chloranyl-3-methyl-1-benzothiophene-2-sulfonamide | | 著者 | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | 登録日 | 2019-09-04 | | 公開日 | 2020-04-22 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
1OIP
 
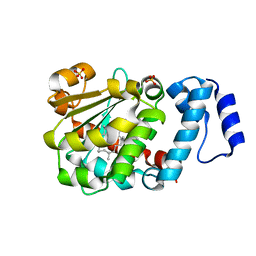 | | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein | | 分子名称: | (2R)-2,5,7,8-TETRAMETHYL-2-[(4R,8R)-4,8,12-TRIMETHYLTRIDECYL]CHROMAN-6-OL, ALPHA-TOCOPHEROL TRANSFER PROTEIN, SULFATE ION | | 著者 | Meier, R, Tomizaki, T, Schulze-Briese, C, Baumann, U, Stocker, A. | | 登録日 | 2003-06-24 | | 公開日 | 2004-01-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Molecular Basis of Vitamin E Retention: Structure of Human Alpha-Tocopherol Transfer Protein
J.Mol.Biol., 331, 2003
|
|
5KB1
 
 | | Crystal Structure of a Tris-thiolate Hg(II) Complex in a de Novo Three Stranded Coiled Coil Peptide | | 分子名称: | CHLORIDE ION, Hg(II)Zn(II)(GRAND Coil Ser-L16CL30H)3+, MERCURY (II) ION, ... | | 著者 | Ruckcthong, L, Zastrow, M.L, Stuckey, J.A, Pecoraro, V.L. | | 登録日 | 2016-06-02 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | A Crystallographic Examination of Predisposition versus Preorganization in de Novo Designed Metalloproteins.
J.Am.Chem.Soc., 138, 2016
|
|
6P5R
 
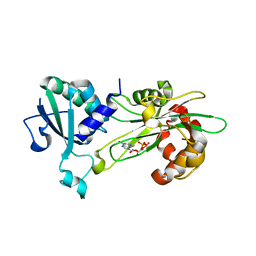 | | Structure of T. brucei MERS1-GDP complex | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Mitochondrial edited mRNA stability factor 1 | | 著者 | Schumacher, M.A. | | 登録日 | 2019-05-30 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
4Z1W
 
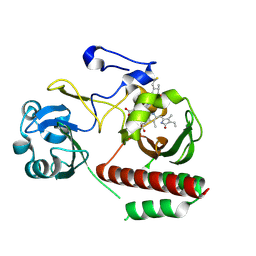 | | CRYSTAL STRUCTURE OF MONOMERIC BACTERIOPHYTOCHROME mutant D207L Y263F From Synchrotron | | 分子名称: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, 3-[2-[(Z)-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E,4R)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome | | 著者 | Bhattacharya, S, Satyshur, K.A, Wangkanont, K, Lehtivuori, H, Forest, K.T. | | 登録日 | 2015-03-27 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Removal of Chromophore-Proximal Polar Atoms Decreases Water Content and Increases Fluorescence in a Near Infrared Phytofluor.
Front Mol Biosci, 2, 2015
|
|
8POV
 
 | | Crystal Structure of the C19G/C120G variant of the membrane-bound [NiFe]-Hydrogenase from Cupriavidus necator in the H2-reduced state at 1.92 A Resolution. | | 分子名称: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | 著者 | Schmidt, A, Kalms, J, Scheerer, P. | | 登録日 | 2023-07-05 | | 公開日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Stepwise conversion of the Cys 6 [4Fe-3S] to a Cys 4 [4Fe-4S] cluster and its impact on the oxygen tolerance of [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
6SCE
 
 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | 分子名称: | Uncharacterized protein, cyclic oligoadenylate | | 著者 | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | 登録日 | 2019-07-24 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
1KZZ
 
 | | DOWNSTREAM REGULATOR TANK BINDS TO THE CD40 RECOGNITION SITE ON TRAF3 | | 分子名称: | TNF receptor associated factor 3, TRAF family member-associated NF-kappa-b activator | | 著者 | Li, C, Ni, C.-Z, Havert, M.L, Cabezas, E, He, J, Kaiser, D, Reed, J.C, Satterthwait, A.C, Cheng, G, Ely, K.R. | | 登録日 | 2002-02-08 | | 公開日 | 2002-04-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Downstream regulator TANK binds to the CD40 recognition site on TRAF3.
Structure, 10, 2002
|
|
6VBM
 
 | | Crystal structure of a S310A mutant of PBP2 from Neisseria gonorrhoeae | | 分子名称: | PHOSPHATE ION, Probable peptidoglycan D,D-transpeptidase PenA | | 著者 | Singh, A, Davies, C. | | 登録日 | 2019-12-19 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Mutations in Neisseria gonorrhoeae penicillin-binding protein 2 associated with extended-spectrum cephalosporin resistance create an energetic barrier against acylation via restriction of protein dynamics
J.Biol.Chem., 2020
|
|
5JOY
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraLOG | | 分子名称: | (Z)-L-Arabinonhydroximo-1,4-lactone, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | 登録日 | 2016-05-03 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | 分子名称: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-29 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | 分子名称: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | 著者 | Kovalevsky, A, Kneller, D.W, Coates, L. | | 登録日 | 2021-07-26 | | 公開日 | 2021-11-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
