4XC3
 
 | | Crystal structure of human 4E10 Fab in complex with its peptide epitope on HIV-1 gp41; crystals cryoprotected with rac-glycerol 1-phosphate | | Descriptor: | 4E10 Fab heavy chain, 4E10 Fab light chain, MODIFIED FRAGMENT OF HIV-1 GLYCOPROTEIN (GP41) INCLUDING THE MPER REGION 671-683, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-17 | | Release date: | 2016-02-03 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
6RU0
 
 | |
4XCY
 
 | | Crystal structure of human 4E10 Fab in complex with phosphatidylglycerol (06:0 PG) | | Descriptor: | (2S)-3-{[(R)-{[(2R)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-2-(hexanoyloxy)propyl hexanoate, 4E10 Fab heavy chain, 4E10 Fab light chain, ... | | Authors: | Irimia, A, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10.
Immunity, 44, 2016
|
|
7A2K
 
 | |
1AAX
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1A0G
 
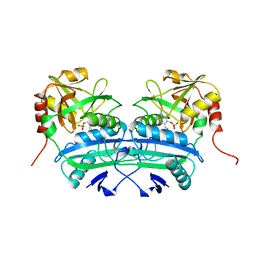 | | L201A MUTANT OF D-AMINO ACID AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAMINE-5'-PHOSPHATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, D-AMINO ACID AMINOTRANSFERASE | | Authors: | Sugio, S, Kashima, A, Kishimoto, K, Peisach, D, Petsko, G.A, Ringe, D, Yoshimura, T, Esaki, N. | | Deposit date: | 1997-11-30 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of L201A mutant of D-amino acid aminotransferase at 2.0 A resolution: implication of the structural role of Leu201 in transamination.
Protein Eng., 11, 1998
|
|
6RVG
 
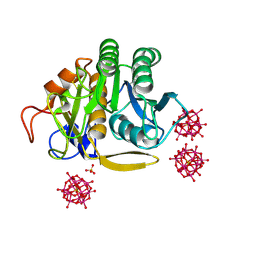 | |
1A5L
 
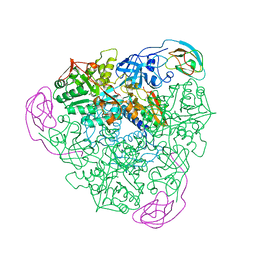 | | K217C VARIANT OF KLEBSIELLA AEROGENES UREASE | | Descriptor: | UREASE (ALPHA SUBUNIT), UREASE (BETA SUBUNIT), UREASE (GAMMA SUBUNIT) | | Authors: | Pearson, M.A, Schaller, R.A, Michel, L.O, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chemical rescue of Klebsiella aerogenes urease variants lacking the carbamylated-lysine nickel ligand.
Biochemistry, 37, 1998
|
|
5FQ6
 
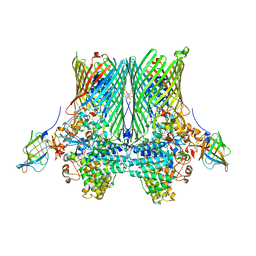 | | Crystal structure of the SusCD complex BT2261-2264 from Bacteroides thetaiotaomicron | | Descriptor: | 3-decanoyloxypropyl decanoate, BT_2261, CALCIUM ION, ... | | Authors: | Glenwright, A.J, Pothula, K.R, Chorev, D.S, Basle, A, Robinson, C.V, Kleinekathoefer, U, Bolam, D.N, van den Berg, B. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nutrient acquisition by dominant members of the human gut microbiota.
Nature, 541, 2017
|
|
6ROH
 
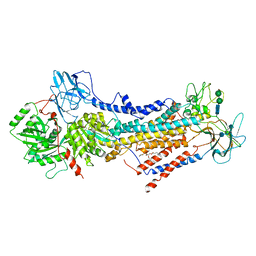 | | Cryo-EM structure of the autoinhibited Drs2p-Cdc50p | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
5MD6
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=-6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
6RVU
 
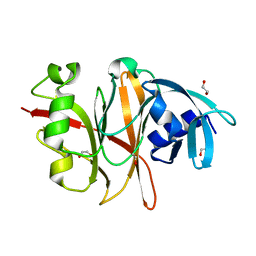 | | Crystal structure of the Burkholderia Lethal Factor 1 (BLF1) | | Descriptor: | 1,2-ETHANEDIOL, Lethal Factor 1 (BLF1) | | Authors: | Mobbs, G.W, Aziz, A.A, Blackburn, G.M, Sedelnikova, S.E, Minshull, T.C, Dickman, M.J, Baker, P.J, Nathan, S, Firdaus-Raih, M, Rice, D.W. | | Deposit date: | 2019-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Molecular basis of specificity and deamidation of eIF4A by Burkholderia Lethal Factor 1.
Commun Biol, 5, 2022
|
|
6EJZ
 
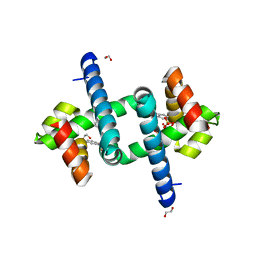 | | Tryptophan Repressor TrpR from E.coli variant S88Y with Indole-3-acetic acid as ligand | | Descriptor: | 1,2-ETHANEDIOL, 1H-INDOL-3-YLACETIC ACID, SULFATE ION, ... | | Authors: | Stiel, A.C, Shanmugaratnam, S, Herud-Sikimic, O, Juergens, G, Hocker, B. | | Deposit date: | 2017-09-24 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A biosensor for the direct visualization of auxin
Nature, 2021
|
|
6AM9
 
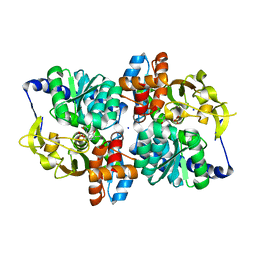 | | Engineered tryptophan synthase b-subunit from Pyrococcus furiosus, PfTrpB2B9, with Ser-bound in a predominantly closed state. | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, SODIUM ION, Tryptophan synthase beta chain 1, ... | | Authors: | Buller, A.R, van Roye, P. | | Deposit date: | 2017-08-09 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
J. Am. Chem. Soc., 140, 2018
|
|
3LQ3
 
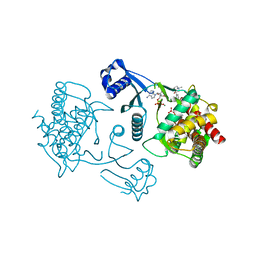 | | Crystal structure of human choline kinase beta in complex with phosphorylated hemicholinium-3 and adenosine nucleotide | | Descriptor: | (2S)-2-[4'-({dimethyl[2-(phosphonooxy)ethyl]ammonio}acetyl)biphenyl-4-yl]-2-hydroxy-4,4-dimethylmorpholin-4-ium, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
5G1A
 
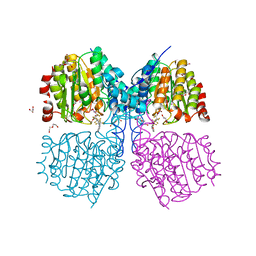 | | Bordetella Alcaligenes HDAH bound to PFSAHA | | Descriptor: | 2,2,3,3,4,4,5,5,6,6,7,7-dodecakis(fluoranyl)-~{N}-oxidanyl-~{N}'-phenyl-octanediamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
5MLW
 
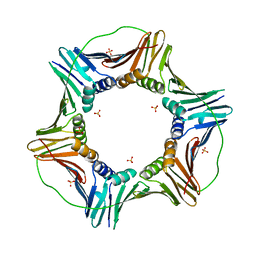 | |
7R1Q
 
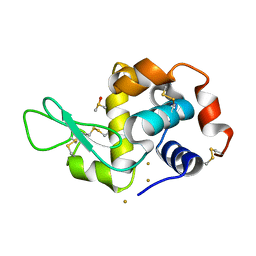 | |
5J6Q
 
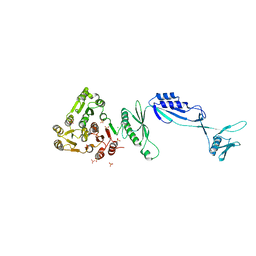 | | Cwp8 from Clostridium difficile | | Descriptor: | CHLORIDE ION, Cell wall binding protein cwp8, SULFATE ION | | Authors: | Renko, M, Usenik, A, Turk, D. | | Deposit date: | 2016-04-05 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The CWB2 Cell Wall-Anchoring Module Is Revealed by the Crystal Structures of the Clostridium difficile Cell Wall Proteins Cwp8 and Cwp6.
Structure, 25, 2017
|
|
8QPW
 
 | |
6X97
 
 | |
6EQC
 
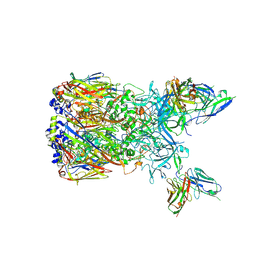 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
7A2Q
 
 | |
8WOW
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, I160L/G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, UDP-glucose 4-epimerase 2, ... | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8B5U
 
 | |
