2CWC
 
 | |
6NS5
 
 | | Crystal structure of fungal lipoxygenase from Fusarium graminearum. Second C2 crystal form. | | Descriptor: | FE (II) ION, lipoxygenase | | Authors: | Pakhomova, S, Boeglin, W.E, Neau, D.B, Bartlett, S.G, Brash, A.R, Newcomer, M.E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | An ensemble of lipoxygenase structures reveals novel conformations of the Fe coordination sphere.
Protein Sci., 28, 2019
|
|
2CXY
 
 | |
7ZL4
 
 | | Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii | | Descriptor: | CsuA/B | | Authors: | Pakharukova, N, Malmi, H, Tuittila, M, Paavilainen, S, Ghosal, D, Chang, Y.W, Jensen, G.J, Zavialov, A.V. | | Deposit date: | 2022-04-13 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Archaic chaperone-usher pili self-secrete into superelastic zigzag springs.
Nature, 609, 2022
|
|
1CC7
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
2M1I
 
 | |
1CDS
 
 | | STRUCTURE OF A SOLUBLE, GLYCOSYLATED FORM OF THE HUMAN COMPLEMENT REGULATORY PROTEIN CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 | | Authors: | Fletcher, C.M, Harrison, R.A, Lachmann, P.J, Neuhaus, D. | | Deposit date: | 1994-06-01 | | Release date: | 1994-09-30 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure of a soluble, glycosylated form of the human complement regulatory protein CD59.
Structure, 2, 1994
|
|
6WFS
 
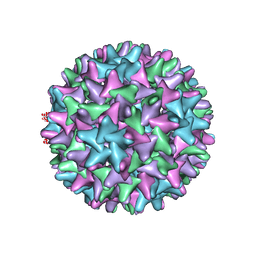 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | Descriptor: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | Authors: | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
2M1L
 
 | | Solution NMR Structure of Cyclin-dependent kinase 2-associated protein 2 (CDK2AP2, DOC-1R) from Homo sapiens, Northeast Structural Genomics Consortium (NESG) Target HR8910C | | Descriptor: | Cyclin-dependent kinase 2-associated protein 2 | | Authors: | Ertekin, A, Janjua, H, Kohan, E, Shastry, R, Pederson, K, Prestegard, J.H, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of CDK2-associated protein 2 (CDK2AP2, Deleted in Oral Cancer 1 Related protein, DOC-1R)
To be Published
|
|
8OYZ
 
 | |
4X4J
 
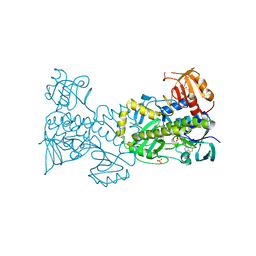 | | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative oxygenase, SULFATE ION | | Authors: | Tsai, S.-C, Jackson, D.R, Patel, A, Barajas, J.F, Rohr, J, Yu, X, Liu, H.-W, Sasaki, E, Calveras, J, Metsa-Ketela, M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural and Functional Studies of BexE: Insights into Oxidation During BE-7585A Biosynthesis
To Be Published
|
|
1CBO
 
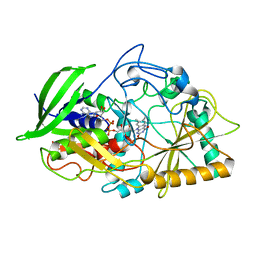 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES HIS447ASN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-02-26 | | Release date: | 1999-03-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
2D9F
 
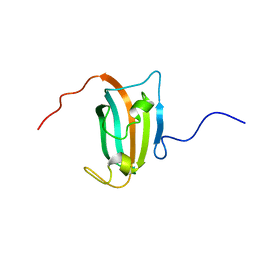 | | Solution structure of RUH-047, an FKBP domain from human cDNA | | Descriptor: | FK506-binding protein 8 variant | | Authors: | Ruhul Momen, A.Z.M, Hirota, H, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-09 | | Release date: | 2006-06-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RUH-047, an FKBP domain from human cDNA
To be Published
|
|
7P65
 
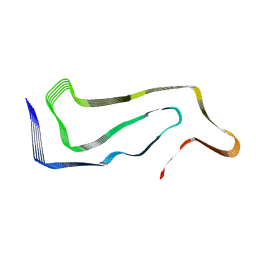 | | Progressive supranuclear palsy tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
5HBD
 
 | | Filamentous Assembly of Green Fluorescent Protein Supported by a C-terminal fusion of 18-residues, viewed in space group C2 | | Descriptor: | Green fluorescent protein | | Authors: | Sawaya, M.R, Hochschild, A, Heller, D.M, McPartland, L, Eisenberg, D.S. | | Deposit date: | 2015-12-31 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Green Fluorescent Protein Fusion that Self Assembles as Polar Filaments
to be published
|
|
6THQ
 
 | | Crystal structure of branched-chain aminotransferase from thermophilic archaea Thermoproteus uzoniensis with norvaline | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Bezsudnova, E.Y, Popov, V.O. | | Deposit date: | 2019-11-21 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-Dimensional Structure of Branched-Chain Amino Acid Transaminase from Thermoproteus uzoniensis in Complex with L-Norvaline
Crystallography Reports, 2020
|
|
7P66
 
 | | Globular glial tauopathy type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P68
 
 | | Globular glial tauopathy type 3 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P67
 
 | | Globular glial tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
7P6C
 
 | | Limbic-predominant neuronal inclusion body 4R tauopathy type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
4PE6
 
 | | Crystal structure of ABC transporter solute binding protein from Thermobispora bispora DSM 43833 | | Descriptor: | (2R,3S)-2,3,4-trihydroxybutanoic acid, Putative ABC transporter | | Authors: | Chang, C, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of ABC transporter solute binding protein from Thermobispora bispora DSM 43833
to be published
|
|
7P6E
 
 | | Argyrophilic grain disease type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
8F9Y
 
 | | SAL1 from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, SAL1 phosphatase | | Authors: | Frkic, R.L, Kaczmarski, J.A, Tan, L, Jackson, C.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sensing and signaling of oxidative stress in chloroplasts by inactivation of the SAL1 phosphoadenosine phosphatase.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7P6D
 
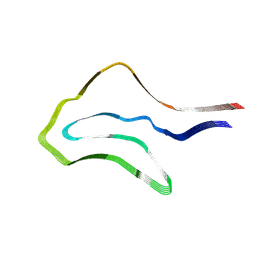 | | Argyrophilic grain disease type 1 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
