7OOU
 
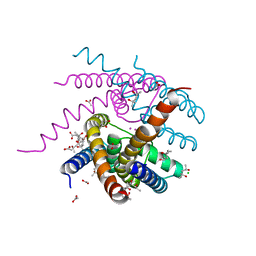 | | NaK C-DI mutant with Li+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
4U0K
 
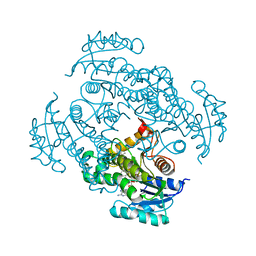 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase complexed with N-(5-chloro-2-methylphenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide | | Descriptor: | (3S)-N-(5-CHLORO-2-METHYLPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
4U0W
 
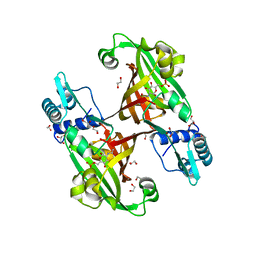 | |
7OQ1
 
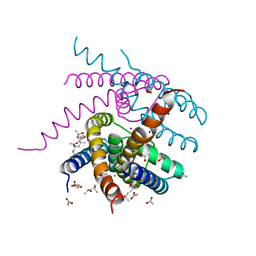 | | NaK S-ELM mutant with Na+ and K+ | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
4U3V
 
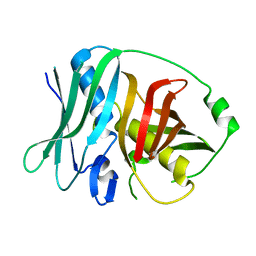 | |
4U48
 
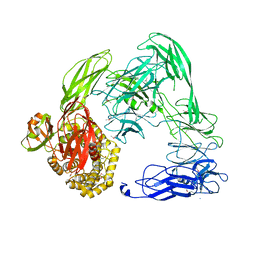 | |
4U4J
 
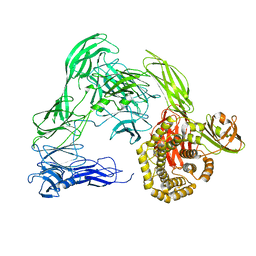 | |
6XM3
 
 | | Structure of SARS-CoV-2 spike at pH 5.5, single RBD up, conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhou, T, Tsybovsky, Y, Olia, A, Kwong, P.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-08-12 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
4UE3
 
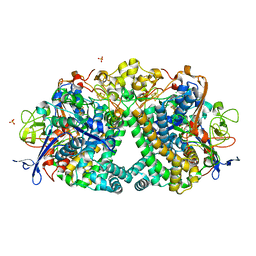 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
7OV9
 
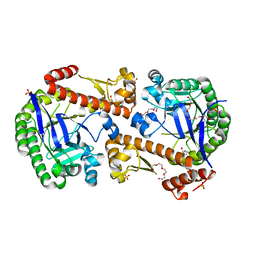 | | Heterodimeric tRNA-Guanine Transglycosylase from mouse, apo-structure | | Descriptor: | HEXAETHYLENE GLYCOL, Queuine tRNA-ribosyltransferase accessory subunit 2, Queuine tRNA-ribosyltransferase catalytic subunit 1, ... | | Authors: | Sebastiani, M, Heine, A, Reuter, K. | | Deposit date: | 2021-06-14 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Investigation of the Heterodimeric Murine tRNA-Guanine Transglycosylase.
Acs Chem.Biol., 17, 2022
|
|
6XU1
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with GTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
4U5Y
 
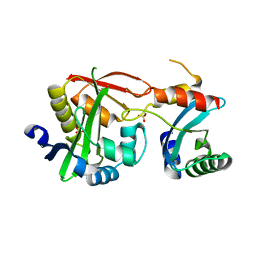 | | Crystal Structure of the complex between the GNAT domain of S. lividans PAT and the acetyl-CoA synthetase C-terminal domain of S. enterica | | Descriptor: | Acetyl-coenzyme A synthetase, Acyl-CoA synthetase, SULFATE ION | | Authors: | Taylor, K.C, Tucker, A.C, Rank, K.C, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2014-07-25 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Insights into the specificity of lysine acetyltransferases.
J.Biol.Chem., 289, 2014
|
|
4U6Z
 
 | |
5LZA
 
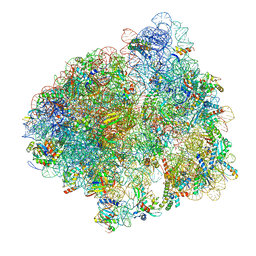 | | Structure of the 70S ribosome with SECIS-mRNA and P-site tRNA (Initial complex, IC) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
8BIP
 
 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-02 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6XHW
 
 | | Crystal structure of the A2058-unmethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
4U8D
 
 | |
6OD9
 
 | | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, CESIUM ION, MAGNESIUM ION, ... | | Authors: | Jones, C.P, Tran, B, Ferre-D'Amare, A.R. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.102 Å) | | Cite: | Co-crystal structure of the Fusobacterium ulcerans ZTP riboswitch using an X-ray free-electron laser.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8BJQ
 
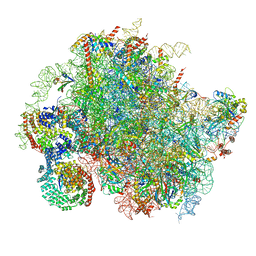 | | Structure of a yeast 80S ribosome-bound N-Acetyltransferase B complex | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Knorr, A.G, Mackens-Kiani, T, Musial, J, Berninghausen, O, Becker, T, Beatrix, B, Beckmann, R. | | Deposit date: | 2022-11-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The dynamic architecture of Map1- and NatB-ribosome complexes coordinates the sequential modifications of nascent polypeptide chains.
Plos Biol., 21, 2023
|
|
6X9D
 
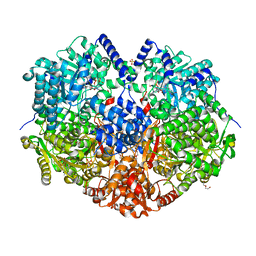 | |
5LZF
 
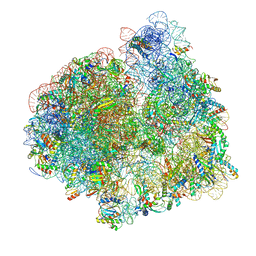 | | Structure of the 70S ribosome with fMetSec-tRNASec in the hybrid pre-translocation state (H) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Fischer, N, Neumann, P, Bock, L.V, Maracci, C, Wang, Z, Paleskava, A, Konevega, A.L, Schroeder, G.F, Grubmueller, H, Ficner, R, Rodnina, M.V, Stark, H. | | Deposit date: | 2016-09-29 | | Release date: | 2016-11-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | The pathway to GTPase activation of elongation factor SelB on the ribosome.
Nature, 540, 2016
|
|
6XZ4
 
 | | Crystal structure of TLNRD1 | | Descriptor: | Talin rod domain-containing protein 1 | | Authors: | Cowell, A, Singh, A.K, Brown, D.G, Goult, B.T. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Talin rod domain-containing protein 1 (TLNRD1) is a novel actin-bundling protein which promotes filopodia formation.
J.Cell Biol., 220, 2021
|
|
6XWG
 
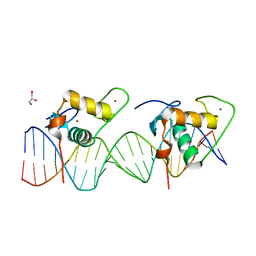 | | Crystal Structure of the Human RXR/RAR DNA-Binding Domain Heterodimer Bound to the Human RARb2 DR5 Response Element | | Descriptor: | CHLORIDE ION, GLYCEROL, RARb2 DR5 Response Element, ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Peluso-Iltis, C, Rochel, N. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for DNA recognition and allosteric control of the retinoic acid receptors RAR-RXR.
Nucleic Acids Res., 48, 2020
|
|
4UAN
 
 | |
4UCK
 
 | | X-ray structure and activities of an essential Mononegavirales L- protein domain | | Descriptor: | RNA-DIRECTED RNA POLYMERASE L, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Paesen, G.C, Collet, A, Sallamand, C, Debart, F, Vasseur, J.J, Canard, B, Decroly, E, Grimes, J.M. | | Deposit date: | 2014-12-03 | | Release date: | 2015-11-18 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure and Activities of an Essential Mononegavirales L-Protein Domain.
Nat.Commun., 6, 2015
|
|
