5VBG
 
 | |
5UPP
 
 | | Crystal structure of human fumarate hydratase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fumarate hydratase, mitochondrial | | Authors: | Rangel, V.L, Ajalla, M.A, Rustiguel, J.K, Pereira de Padua, R.A, Nonato, M.C. | | Deposit date: | 2017-02-03 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, biochemical and biophysical characterization of recombinant human fumarate hydratase.
Febs J., 2019
|
|
6CQG
 
 | |
6CHX
 
 | |
5UQF
 
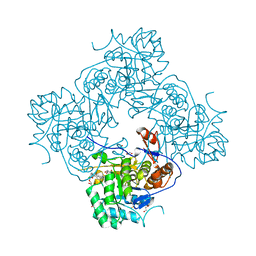 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with IMP and the inhibitor P225 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Campylobacter jejuni in the complex with IMP and the inhibitor P225
To Be Published
|
|
4V3J
 
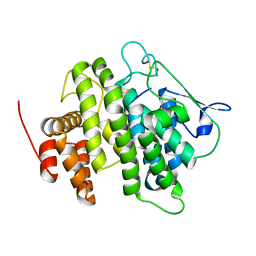 | | Structural and functional characterization of a novel monotreme- specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MONOTREME LACTATING PROTEIN | | Authors: | Kumar, A, Newman, J, Polekina, G, Adams, T.E, Sharp, J.A, Peat, T.S, Nicholas, K.R. | | Deposit date: | 2014-10-20 | | Release date: | 2016-01-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6CJO
 
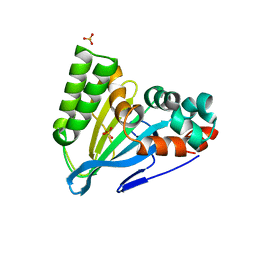 | | Crystal Structure of Chalcone Isomerase from Medicago Sativa with the G95S mutation. | | Descriptor: | Chalcone--flavonone isomerase 1, SULFATE ION | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6CLF
 
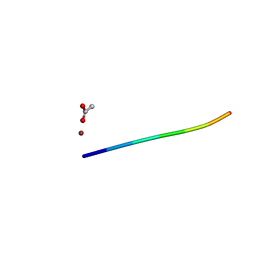 | | 1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLO
 
 | | 1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CW1
 
 | |
4V9F
 
 | |
6CXU
 
 | | Structure of the S167H mutant of human indoleamine 2,3 dioxygenase in complex with tryptophan and cyanide | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.-R, Karkashon, S, Batabyal, D, Poulos, T.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
4V05
 
 | | FGFR1 in complex with AZD4547. | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V67
 
 | | Crystal structure of a translation termination complex formed with release factor RF2. | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Asahara, H, Lancaster, L, Laurberg, M, Hirschi, A, Noller, H.F. | | Deposit date: | 2008-10-27 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a translation termination complex formed with release factor RF2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6CQK
 
 | | Crystal Structure of mitochondrial single-stranded DNA binding proteins from S. cerevisiae, Rim1 (Form1) | | Descriptor: | SsDNA-binding protein essential for mitochondrial genome maintenance | | Authors: | Singh, S.P, Kukshal, V, Bona, P.D, Lytle, A.K, Edwin, A, Galletto, R. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mitochondrial single-stranded DNA binding protein from S. cerevisiae, Rim1, does not form stable homo-tetramers and binds DNA as a dimer of dimers.
Nucleic Acids Res., 46, 2018
|
|
6CRV
 
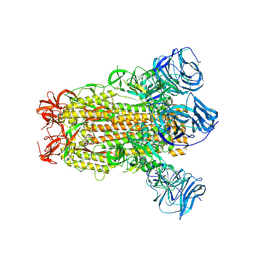 | | SARS Spike Glycoprotein, Stabilized variant, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | Authors: | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-04-11 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
4UYQ
 
 | |
2VTS
 
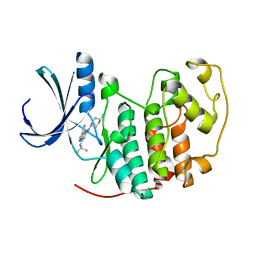 | | Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H- pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design. | | Descriptor: | 5-[(4-AMINOCYCLOHEXYL)AMINO]-7-(PROPAN-2-YLAMINO)PYRAZOLO[1,5-A]PYRIMIDINE-3-CARBONITRILE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wyatt, P.G, Woodhead, A.J, Boulstridge, J.A, Berdini, V, Carr, M.G, Cross, D.M, Danillon, D, Davis, D.J, Devine, L.A, Early, T.R, Feltell, R.E, Lewis, E.J, McMenamin, R.L, Navarro, E.F, O'Brien, M.A, O'Reilly, M, Reule, M, Saxty, G, Seavers, L.C.A, Smith, D, Squires, M.S, Trewartha, G, Walker, M.T, Woolford, A.J. | | Deposit date: | 2008-05-15 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of N-(4-Piperidinyl)-4-(2,6-Dichlorobenzoylamino)-1H-Pyrazole-3-Carboxamide (at7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-Based X-Ray Crystallography and Structure Based Drug Design.
J.Med.Chem., 51, 2008
|
|
6CEA
 
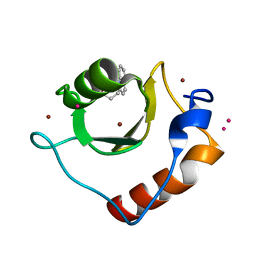 | | Crystal structure of fragment 3-(quinolin-2-yl)propanoic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(quinolin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Ravichandran, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
5V4R
 
 | |
6CEE
 
 | | Crystal structure of fragment 3-(1-Methyl-2-oxo-1,2-dihydroquinoxalin-3-yl)propionic acid bound in the ubiquitin binding pocket of the HDAC6 zinc-finger domain | | Descriptor: | 3-(4-methyl-3-oxo-3,4-dihydroquinoxalin-2-yl)propanoic acid, Histone deacetylase 6, UNKNOWN ATOM OR ION, ... | | Authors: | Harding, R.J, Halabelian, L, Ferreira de Freitas, R, Franzoni, I, Ravichandran, M, Lautens, M, Santhakumar, V, Schapira, M, Bountra, C, Edwards, A.M, Arrowsmith, C.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-11 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Structure-Activity Relationship of HDAC6 Zinc-Finger Ubiquitin Binding Domain Inhibitors.
J. Med. Chem., 61, 2018
|
|
6CC4
 
 | | Structure of MurJ from Escherichia coli | | Descriptor: | PHOSPHATE ION, soluble cytochrome b562, lipid II flippase MurJ chimera | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and mutagenic analysis of the lipid II flippase MurJ fromEscherichia coli.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CCG
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA, Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
5VCB
 
 | |
4V7X
 
 | | Structure of the Thermus thermophilus ribosome complexed with erythromycin. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-17 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
