4XFY
 
 | |
4XG1
 
 | | Psychromonas ingrahamii diaminopimelate decarboxylase with LLP | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, Diaminopimelate decarboxylase, POTASSIUM ION, ... | | Authors: | Peverelli, M.G, Wubben, J.M, Panjikar, S, Perugini, M.A. | | Deposit date: | 2014-12-30 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression to crystallization of diaminopimelate decarboxylase from the psychrophile Psychromonas ingrahamii
To Be Published
|
|
4XPI
 
 | | Fe protein independent substrate reduction by nitrogenase variants altered in intramolecular electron transfer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Danyal, K, Rasmusen, A.J, Keable, S.M, Shaw, S, Zadvornyy, O, Duval, S, Dean, D.R, Raugei, S, Peters, J.W, Seefeldt, L.C. | | Deposit date: | 2015-01-17 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fe protein-independent substrate reduction by nitrogenase MoFe protein variants.
Biochemistry, 54, 2015
|
|
5SWH
 
 | | c-Src V281C kinase domain in complex with Rao-IV-151 | | Descriptor: | (2R)-3-[4-amino-5-(4-chlorophenyl)-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl]-2-cyano-N-(propan-2-yl)propanami de, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Merritt, E.A, Dieter, E.M. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A combined approach reveals a regulatory mechanism coupling Src's kinase activity, localization, and phosphotransferase-independent functions
Mol.Cell, 2019
|
|
6HS9
 
 | |
8T6N
 
 | | Crystal structure of T33-27.1: Deep-learning sequence design of co-assembling tetrahedron protein nanoparticles | | Descriptor: | T33-27.1 : A, T33-27.1 : B | | Authors: | Bera, A.K, de Haas, R.J, Kang, A, Sankaran, B, King, N.P. | | Deposit date: | 2023-06-16 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Rapid and automated design of two-component protein nanomaterials using ProteinMPNN.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6WJE
 
 | | Copper resistance protein copG- Form 2 | | Descriptor: | ACETATE ION, COPPER (II) ION, DUF411 domain-containing protein, ... | | Authors: | Hausrath, A.C, Ly, A.T, McEvoy, M.M. | | Deposit date: | 2020-04-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The bacterial copper resistance protein CopG contains a cysteine-bridged tetranuclear copper cluster.
J.Biol.Chem., 295, 2020
|
|
4XUR
 
 | |
8SIS
 
 | |
3RMM
 
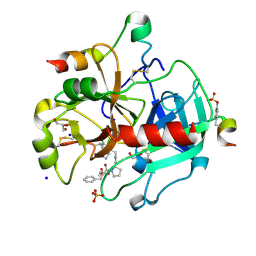 | | Human Thrombin in complex with MI332 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hirudin variant-2, ... | | Authors: | Biela, A, Heine, A, Klebe, G. | | Deposit date: | 2011-04-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect
J.Med.Chem., 55, 2012
|
|
8PTQ
 
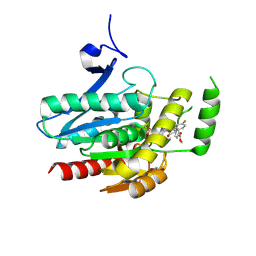 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5l | | Descriptor: | 1,2-ETHANEDIOL, Monoglyceride lipase, methyl 4-[(2~{S},3~{R})-3-(4-fluorophenyl)-1-(1-methanoylpiperidin-4-yl)-4-oxidanylidene-azetidin-2-yl]benzoate | | Authors: | Butini, S, Grether, U, Benz, J, Leibrock, L, Maramai, S, Papa, A, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
5IWO
 
 | | Bacterial sodium channel pore domain, low bromide | | Descriptor: | BROMIDE ION, Ion transport protein | | Authors: | Shaya, D, Findeisen, F, Rohaim, A, Minor, D.L. | | Deposit date: | 2016-03-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Unfolding of a Temperature-Sensitive Domain Controls Voltage-Gated Channel Activation.
Cell, 164, 2016
|
|
6HIZ
 
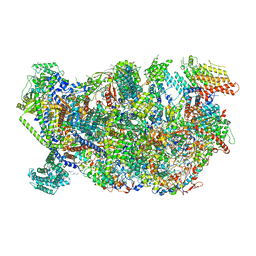 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the head of the small mitoribosomal subunit | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (143-MER), ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boehringer, A, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
7T69
 
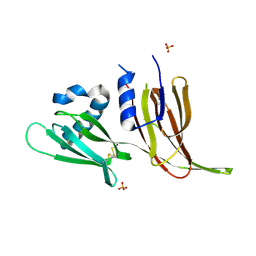 | | Crystal structure of Avr3 (SIX1) from Fusarium oxysporum f. sp. lycopersici | | Descriptor: | Avr3 (SIX1), Secreted in xylem 1, SULFATE ION | | Authors: | Yu, D.S, Outram, M.A, Ericsson, D.J, Jones, D.A, Williams, S.J. | | Deposit date: | 2021-12-13 | | Release date: | 2023-01-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structural repertoire of Fusarium oxysporum f. sp. lycopersici effectors revealed by experimental and computational studies
Elife, 2023
|
|
2X11
 
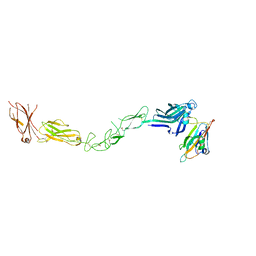 | | Crystal structure of the complete EphA2 ectodomain in complex with ephrin A5 receptor binding domain | | Descriptor: | EPHRIN TYPE-A RECEPTOR 2, EPHRIN-A5 | | Authors: | Seiradake, E, Harlos, K, Sutton, G, Aricescu, A.R, Jones, E.Y. | | Deposit date: | 2009-12-21 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.83 Å) | | Cite: | An Extracellular Steric Seeding Mechanism for Eph-Ephrin Signalling Platform Assembly
Nat.Struct.Mol.Biol., 17, 2010
|
|
6YXW
 
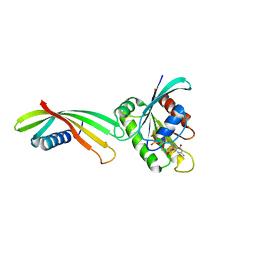 | | Affimer K3 - KRAS protein complex | | Descriptor: | Affimer K3, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Trinh, C.H, Haza, K.Z, Rao, A, Martin, H.L, Tiede, C, Edwards, T.A, McPherson, M.J, Tomlinson, D.C. | | Deposit date: | 2020-05-04 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | RAS-inhibiting biologics identify and probe druggable pockets including an SII-alpha 3 allosteric site.
Nat Commun, 12, 2021
|
|
6LF7
 
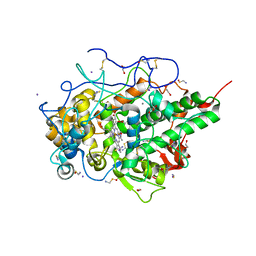 | | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Tyagi, T.K, Singh, R.P, Singh, A.K, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-11-30 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Crystal structure of the complex of goat lactoperoxidase with hypothiocyanite and hydrogen peroxide at 1.79 A resolution.
To Be Published
|
|
3VIK
 
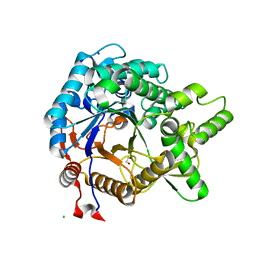 | | Crystal structure of beta-glucosidase from termite Neotermes koshunensis in complex with cellobiose | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucosidase, CHLORIDE ION, ... | | Authors: | Jeng, W.Y, Liu, C.I, Wang, A.H.J. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structures of Neotermes koshunensis beta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6O3L
 
 | |
8ARF
 
 | |
4M0G
 
 | | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor. | | Descriptor: | Adenylosuccinate synthetase, CHLORIDE ION | | Authors: | Tan, K, Zhou, M, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-14 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | The crystal structure of an adenylosuccinate synthetase from Bacillus anthracis str. Ames Ancestor.
To be Published
|
|
4Y0Z
 
 | | Trypsin in complex with with BPTI mutant AMINOBUTYRIC ACID | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Loll, B, Ye, S, Berger, A.A, Muelow, U, Alings, C, Wahl, M.C, Koksch, B. | | Deposit date: | 2015-02-06 | | Release date: | 2015-06-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fluorine teams up with water to restore inhibitor activity to mutant BPTI.
Chem Sci, 6, 2015
|
|
7QNC
 
 | | Cryo-EM structure of human full-length extrasynaptic alpha4beta3delta GABA(A)R in complex with THIP (gaboxadol), histamine and nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,5,6,7-tetrahydro-[1,2]oxazolo[5,4-c]pyridin-3-one, ... | | Authors: | Sente, A, Desai, R, Naydenova, K, Malinauskas, T, Jounaidi, Y, Miehling, J, Zhou, X, Masiulis, S, Hardwick, S.W, Chirgadze, D.Y, Miller, K.W, Aricescu, A.R. | | Deposit date: | 2021-12-20 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Differential assembly diversifies GABA A receptor structures and signalling.
Nature, 604, 2022
|
|
6RGZ
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
3RF1
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, GLYCEROL, Glycyl-tRNA synthetase alpha subunit | | Authors: | Tan, K, Zhang, R, Zhou, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-05 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
