7S4V
 
 | | Cas9 bound to 12-14MM DNA, 60 min time-point, kinked conformation | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, NTS, TS, ... | | Authors: | Bravo, J.P.K, Taylor, D.W, Liu, M.S, Johnson, K.A. | | Deposit date: | 2021-09-09 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for mismatch surveillance by CRISPR-Cas9.
Nature, 603, 2022
|
|
6FJQ
 
 | | Adenovirus species 48, fiber knob protein | | Descriptor: | 1,2-ETHANEDIOL, Fiber, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-01-22 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
4S2X
 
 | | Structure of E. coli RppH bound to RNA and two magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(APC)*GP*U)-3'), RNA pyrophosphohydrolase, ... | | Authors: | Vasilyev, N, Serganov, A. | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of RNA Complexes with the Escherichia coli RNA Pyrophosphohydrolase RppH Unveil the Basis for Specific 5'-End-dependent mRNA Decay.
J.Biol.Chem., 290, 2015
|
|
6VTC
 
 | | p53-specific T cell receptor | | Descriptor: | T-cell Receptor 1a2, p53-specific T cell receptor, B-chain | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-12 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
4YNO
 
 | |
6HI7
 
 | | The ATAD2 bromodomain in complex with compound 10 | | Descriptor: | (2~{R})-~{N}-[5-(3-aminophenyl)-4-ethanoyl-1,3-thiazol-2-yl]-2-azanyl-propanamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.743 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
7NM8
 
 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM, in complex with NADPH | | Descriptor: | Antimycin pathway standalone ketoreductase, AntM, GLYCEROL, ... | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
6HIC
 
 | | The ATAD2 bromodomain in complex with compound 15 | | Descriptor: | (2~{R})-~{N}-[4-ethanoyl-5-[4-(2-oxidanylidenepyrrolidin-1-yl)phenyl]-1,3-thiazol-2-yl]piperazine-2-carboxamide, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Sledz, P, Caflisch, A. | | Deposit date: | 2018-08-29 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Hitting a Moving Target: Simulation and Crystallography Study of ATAD2 Bromodomain Blockers.
Acs Med.Chem.Lett., 11, 2020
|
|
8SIO
 
 | | Crystal structure of PRMT3 with YD1-66 | | Descriptor: | 5'-S-{3-[N'-(4'-chloro[1,1'-biphenyl]-3-yl)carbamimidamido]propyl}-5'-thioadenosine, Protein arginine N-methyltransferase 3 | | Authors: | Song, X, Dong, A, Arrowsmith, C.H, Edwards, A.M, Deng, Y, Huang, R, Min, J. | | Deposit date: | 2023-04-16 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PRMT3 with YD1-66
To be published
|
|
6W0B
 
 | | Open-gate KcsA soaked in 2 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.604 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6HIZ
 
 | | Cryo-EM structure of the Trypanosoma brucei mitochondrial ribosome - This entry contains the head of the small mitoribosomal subunit | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (143-MER), ... | | Authors: | Ramrath, D.J.F, Niemann, M, Leibundgut, M, Bieri, P, Prange, C, Horn, E.K, Leitner, A, Boehringer, A, Schneider, A, Ban, N. | | Deposit date: | 2018-08-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes.
Science, 362, 2018
|
|
7KBY
 
 | | Artificial Metalloproteins with Dinuclear Iron Centers | | Descriptor: | ACETATE ION, CYANIDE ION, Streptavidin, ... | | Authors: | Miller, K.R, Follmer, A.H, Jasniewski, A.J, Sabuncu, S, Biswas, S, Albert, T, Hendrich, M.P, Moenne-Loccoz, P, Borovik, A.S. | | Deposit date: | 2020-10-03 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Artificial Metalloproteins with Dinuclear Iron-Hydroxido Centers.
J.Am.Chem.Soc., 143, 2021
|
|
5ET6
 
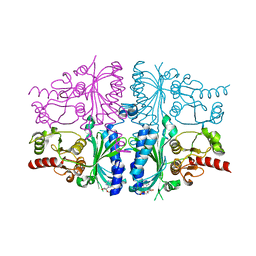 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5L8B
 
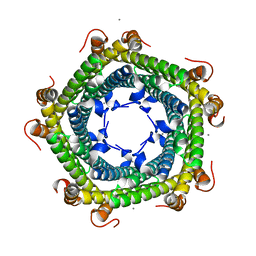 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
6BZH
 
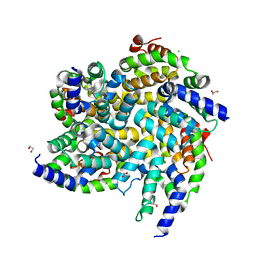 | | Structure of mouse RIG-I tandem CARDs | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Probable ATP-dependent RNA helicase DDX58 | | Authors: | Kershaw, N.J, D'Cruz, A.A, Babon, J.J, Nicholson, S.E. | | Deposit date: | 2017-12-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a second binding site on the TRIM25 B30.2 domain.
Biochem. J., 475, 2018
|
|
5ET7
 
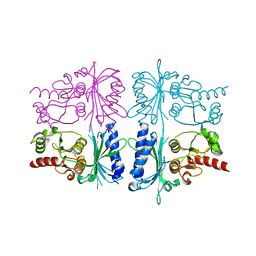 | | Human muscle fructose-1,6-bisphosphatase in inactive T-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2015-11-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | T-to-R switch of muscle fructose-1,6-bisphosphatase involves fundamental changes of secondary and quaternary structure.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
5ETA
 
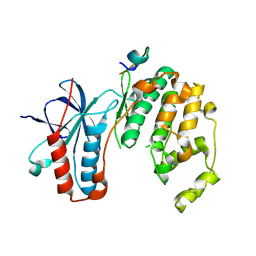 | | Structure of MAPK14 with bound the KIM domain of the Toxoplasma protein GRA24 | | Descriptor: | Mitogen-activated protein kinase 14, Putative transmembrane protein | | Authors: | Pellegrini, E, Palencia, A, Braun, L, Kapp, U, Bougdour, A, Belrhali, H, Bowler, M.W, Hakimi, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-10-26 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Subversion of MAP Kinase Signaling by an Intrinsically Disordered Parasite Secreted Agonist.
Structure, 25, 2017
|
|
7NM7
 
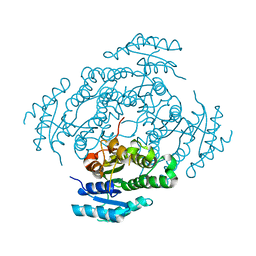 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM | | Descriptor: | Antimycin pathway standalone ketoreductase enzyme, AntM | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
4YQK
 
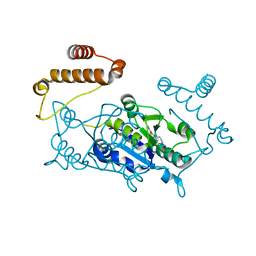 | | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds | | Descriptor: | 4-amino-N-(piperidin-4-yl)-1,2,5-oxadiazole-3-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Elkins, P.A, Bonnette, W.G, Madauss, K.P, Stuckey, J.A. | | Deposit date: | 2015-03-13 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of TrmD, a M1G37 tRNA Methyltransferase with SAM-competitive compounds
To Be Published
|
|
8SPW
 
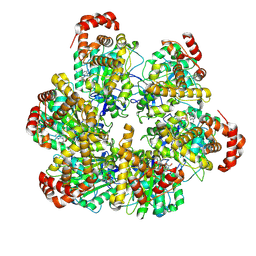 | | PS3 F1 Rotorless, low ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase subunit alpha, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2023-05-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The series of conformational states adopted by rotorless F 1 -ATPase during its hydrolysis cycle.
Structure, 32, 2024
|
|
6FGH
 
 | | Crystal Structure of BAZ2A bromodomain in complex with 3-amino-2-methylpyridine derivative 1 | | Descriptor: | 2-methyl-~{N}-[(2~{R})-1-methylsulfonylpropan-2-yl]pyridin-3-amine, Bromodomain adjacent to zinc finger domain protein 2A | | Authors: | Dalle Vedove, A, Marchand, J.-R, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
5UU5
 
 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8V5C
 
 | | IpaD (122-321) Bound to Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, GLYCEROL, Invasin IpaD | | Authors: | Barker, S.A, Dickenson, N.E, Johnson, S.J. | | Deposit date: | 2023-11-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and functional characterization of the IpaD pi-helix reveals critical roles in DOC interaction, T3SS apparatus maturation, and Shigella virulence.
J.Biol.Chem., 2024
|
|
8KH3
 
 | |
