2J80
 
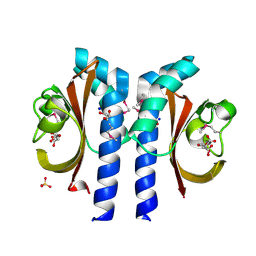 | | Structure of Citrate-bound Periplasmic Domain of Sensor Histidine Kinase CitA | | Descriptor: | CITRATE ANION, GLYCEROL, SENSOR KINASE CITA, ... | | Authors: | Sevvana, M, Vijayan, V, Zweckstetter, M, Reinelt, S, Madden, D.R, Sheldrick, G.M, Bott, M, Griesinger, C, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-10-23 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Ligand-Induced Switch in the Periplasmic Domain of Sensor Histidine Kinase Cita.
J.Mol.Biol., 377, 2008
|
|
2JK4
 
 | | Structure of the human voltage-dependent anion channel | | Descriptor: | VOLTAGE-DEPENDENT ANION-SELECTIVE CHANNEL PROTEIN 1 | | Authors: | Bayrhuber, M, Meins, T, Habeck, M, Becker, S, Giller, K, Villinger, S, Vonrhein, C, Griesinger, C, Zweckstetter, M, Zeth, K. | | Deposit date: | 2008-08-15 | | Release date: | 2008-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of the Human Voltage-Dependent Anion Channel.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2J6D
 
 | | CONKUNITZIN-S2 - CONE SNAIL NEUROTOXIN - DENOVO STRUCTURE | | Descriptor: | CONKUNITZIN-S2 | | Authors: | Korukottu, J, Bayrhuber, M, Montaville, P, Vijayan, V, Jung, Y.-S, Becker, S, Zweckstetter, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-01-16 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | Fast High-Resolution Protein Structure Determination by Using Unassigned NMR Data.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2LYR
 
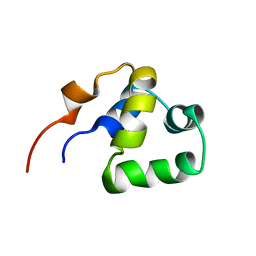 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
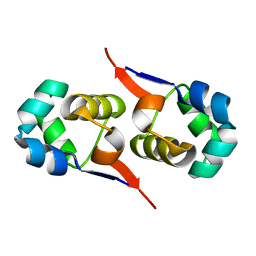 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
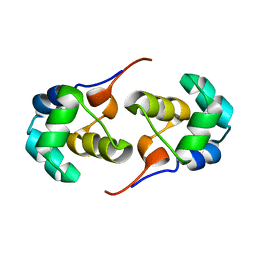 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYS
 
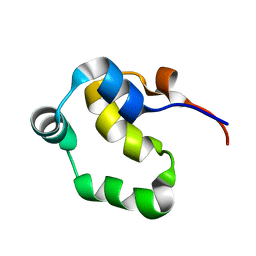 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 257K (-16 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
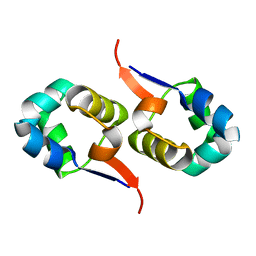 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
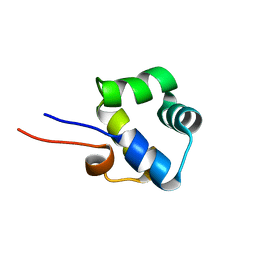 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
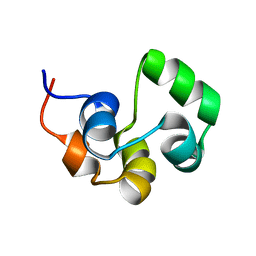 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2N02
 
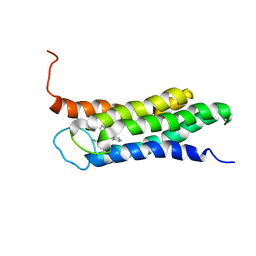 | | Solution structure of the A147T variant of the mitochondrial translocator protein (tspo) in complex with pk11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2015-03-04 | | Release date: | 2015-06-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural Integrity of the A147T Polymorph of Mammalian TSPO.
Chembiochem, 16, 2015
|
|
2MZ7
 
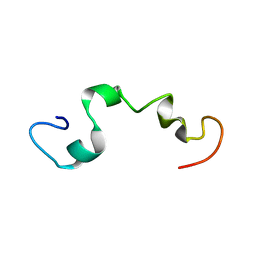 | |
2MF7
 
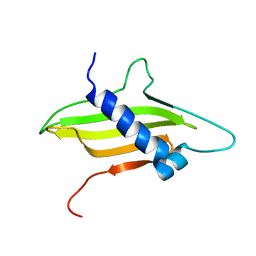 | | Solution structure of the ims domain of the mitochondrial import protein TIM21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bajaj, R, Jaremko, L, Jaremko, M, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis of the Dynamic Structure of the TIM23 Complex in the Mitochondrial Intermembrane Space.
Structure, 22, 2014
|
|
2M1I
 
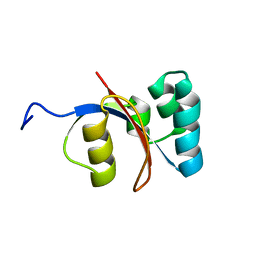 | |
2MGY
 
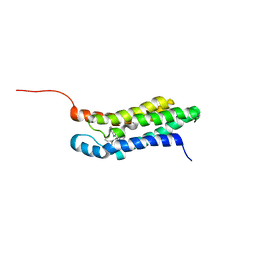 | | Solution structure of the mitochondrial translocator protein (TSPO) in complex with its high-affinity ligand PK11195 | | Descriptor: | N-[(2R)-butan-2-yl]-1-(2-chlorophenyl)-N-methylisoquinoline-3-carboxamide, Translocator protein | | Authors: | Jaremko, M, Jaremko, L, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2013-11-11 | | Release date: | 2014-04-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the mitochondrial translocator protein in complex with a diagnostic ligand.
Science, 343, 2014
|
|
2UVS
 
 | | High Resolution Solid-state NMR structure of Kaliotoxin | | Descriptor: | POTASSIUM CHANNEL TOXIN ALPHA-KTX 3.1 | | Authors: | Korukottu, J, Lange, A, Vijayan, V, Schneider, R, Pongs, O, Becker, S, Baldus, M, Zweckstetter, M. | | Deposit date: | 2007-03-14 | | Release date: | 2008-05-27 | | Last modified: | 2020-01-15 | | Method: | SOLID-STATE NMR | | Cite: | Conformational Plasticity in Ion Channel Recognition of a Peptide Toxin
To be Published
|
|
2GZU
 
 | |
2K3H
 
 | | Structural determinants for Ca2+ and PIP2 binding by the C2A domain of rabphilin-3A | | Descriptor: | CALCIUM ION, Rabphilin-3A | | Authors: | Coudevylle, N, Montaville, P, Leonov, A, Zweckstetter, M, Becker, S. | | Deposit date: | 2008-05-08 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structural Determinants for Ca2+ and Phosphatidylinositol 4,5-Bisphosphate Binding by the C2A Domain of Rabphilin-3A.
J.Biol.Chem., 283, 2008
|
|
2MKC
 
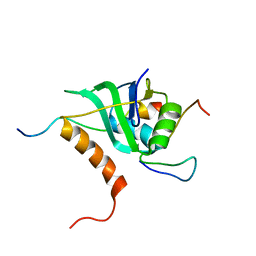 | | Cooperative Structure of the Heterotrimeric pre-mRNA Retention and Splicing Complex | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-splicing factor CWC26, U2 snRNP component IST3 | | Authors: | Wysoczanski, P, Schneider, C, Xiang, S, Munari, F, Trowitzsch, S, Wahl, M.C, Luhrmann, R, Becker, S, Zweckstetter, M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-09-03 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2NBO
 
 | |
2MY3
 
 | |
2NBP
 
 | |
2MY2
 
 | |
4EUI
 
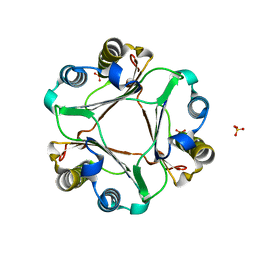 | | Crystal Structure of MIF L46F mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
4ETG
 
 | | Crystal Structure of MIF L46G mutant | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Ashrafi, A, Pojer, F, Lashuel, H. | | Deposit date: | 2012-04-24 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Characterization of molecular determinants of the conformational stability of macrophage migration inhibitory factor: leucine 46 hydrophobic pocket.
Plos One, 7, 2012
|
|
