6WCE
 
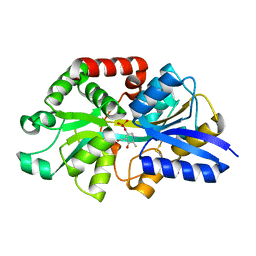 | | Structure of the periplasmic binding protein P5PA | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Pyridoxal-5-phosphate binding protein A (P5PA) | | Authors: | Pan, C, Zimmer, A, Shah, M, Huynh, M, Lai, C.C.L, Sit, B, Hooda, Y, Moraes, T.F. | | Deposit date: | 2020-03-30 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.754 Å) | | Cite: | Actinobacillus utilizes a binding protein-dependent ABC transporter to acquire the active form of vitamin B 6 .
J.Biol.Chem., 297, 2021
|
|
2Q27
 
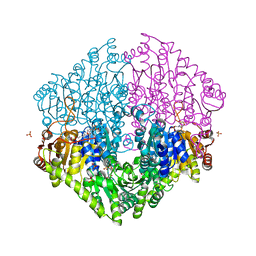 | | Crystal structure of oxalyl-coA decarboxylase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Werther, T, Zimmer, A, Wille, G, Hubner, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli.
Febs J., 277, 2010
|
|
2Q28
 
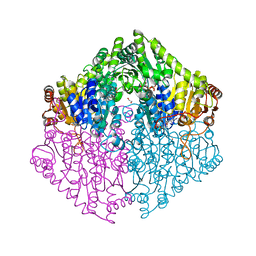 | | Crystal structure of oxalyl-coA decarboxylase from Escherichia coli in complex with adenosine-5`-diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Werther, T, Zimmer, A, Wille, G, Hubner, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli.
Febs J., 277, 2010
|
|
2Q29
 
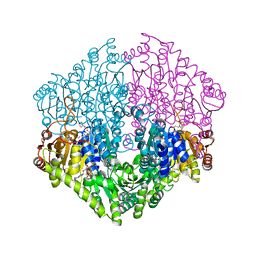 | | Crystal structure of oxalyl-coA decarboxylase from Escherichia coli in complex with acetyl coenzyme A | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYL COENZYME *A, MAGNESIUM ION, ... | | Authors: | Werther, T, Zimmer, A, Wille, G, Hubner, G, Weiss, M.S, Konig, S. | | Deposit date: | 2007-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | New insights into structure-function relationships of oxalyl CoA decarboxylase from Escherichia coli.
Febs J., 277, 2010
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
6ZCI
 
 | | Crystal structure of BRD4-BD1 in complex with NVS-BET-1 | | Descriptor: | (4~{R})-4-(4-chlorophenyl)-1-cyclopropyl-5-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)-3-methyl-4~{H}-pyrrolo[3,4-c]pyrazol-6-one, Bromodomain-containing protein 4 | | Authors: | Faller, M. | | Deposit date: | 2020-06-11 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.976 Å) | | Cite: | BET bromodomain inhibitors regulate keratinocyte plasticity.
Nat.Chem.Biol., 17, 2021
|
|
6TFP
 
 | | BTK in complex with LOU064, a potent and highly selective covalent inhibitor | | Descriptor: | SODIUM ION, Tyrosine-protein kinase BTK, ~{N}-[3-[6-azanyl-5-[2-[methyl(propanoyl)amino]ethoxy]pyrimidin-4-yl]-5-fluoranyl-2-methyl-phenyl]-4-cyclopropyl-2-fluoranyl-benzamide | | Authors: | Scheufler, C, Hinniger, A, Gutmann, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of LOU064 (Remibrutinib), a Potent and Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 63, 2020
|
|
6S90
 
 | | BTK in complex with an inhibitor | | Descriptor: | 4-~{tert}-butyl-~{N}-[2-methyl-3-[6-[4-(4-methylpiperazin-1-yl)carbonylphenyl]-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]phenyl]benzamide, Tyrosine-protein kinase BTK | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2019-07-11 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Design of Potent and Selective Covalent Inhibitors of Bruton's Tyrosine Kinase Targeting an Inactive Conformation.
Acs Med.Chem.Lett., 10, 2019
|
|
4EFU
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 6-Hydroxy-3-(3-methyl-benzyl)-1H-indazole-5-carboxylic acid benzyl-methyl-amide | | Descriptor: | Heat shock protein HSP 90-alpha, N-benzyl-6-hydroxy-N-methyl-3-(3-methylbenzyl)-1H-indazole-5-carboxamide, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EEH
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-(4-Hydroxy-phenyl)-1H-indazol-6-ol | | Descriptor: | 3-(4-hydroxyphenyl)-1H-indazol-6-ol, Heat shock protein HSP 90-alpha, SULFATE ION | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-28 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4EFT
 
 | | Hsp90 Alpha N-terminal Domain in Complex with an Inhibitor 3-Cyclohexyl-2-(6-hydroxy-1H-indazol-3-yl)-propionitrile | | Descriptor: | (2R)-3-cyclohexyl-2-(6-hydroxy-1H-indazol-3-yl)propanenitrile, Heat shock protein HSP 90-alpha | | Authors: | Musil, D, Lehmann, M, Graedler, U, Buchstaller, H.-P. | | Deposit date: | 2012-03-30 | | Release date: | 2012-06-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Fragment-based discovery of hydroxy-indazole-carboxamides as novel small molecule inhibitors of Hsp90
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3TT0
 
 | | Co-structure of Fibroblast Growth Factor Receptor 1 kinase domain with 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (BGJ398) | | Descriptor: | 3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-(6-{[4-(4-ethylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1-methylurea, Basic fibroblast growth factor receptor 1, GLYCEROL, ... | | Authors: | Bussiere, D.E, Murray, J.M, Shu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin-1-yl)-phenylamino]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase.
J.Med.Chem., 54, 2011
|
|
