8KHU
 
 | | Hepatitis B virus core protein Y132A mutant in complex with THPP derivatives 48 | | Descriptor: | (6~{S},7~{R})-6,7-dimethyl-3-(2-oxidanylidenepyrrolidin-1-yl)-~{N}-[3,4,5-tris(fluoranyl)phenyl]-6,7-dihydro-4~{H}-pyrazolo[1,5-a]pyrazine-5-carboxamide, Capsid protein, GLYCEROL, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of 4,5,6,7-Tetrahydropyrazolo[1.5-a]pyrizine Derivatives as Core Protein Allosteric Modulators (CpAMs) for the Inhibition of Hepatitis B Virus.
J.Med.Chem., 66, 2023
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
5XS2
 
 | |
5XQX
 
 | |
8I71
 
 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
7EKN
 
 | | Crystal structure of AF10-ipep complex | | Descriptor: | Protein AF-10, ipep | | Authors: | Chen, S, Zhou, Z. | | Deposit date: | 2021-04-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural characteristics of coiled-coil regions in AF10-DOT1L and AF10-inhibitory peptide complex.
J Leukoc Biol, 110, 2021
|
|
7EDP
 
 | | Crystal structure of AF10-DOT1L complex | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, Protein AF-10 | | Authors: | Chen, S, Zhou, Z. | | Deposit date: | 2021-03-16 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characteristics of coiled-coil regions in AF10-DOT1L and AF10-inhibitory peptide complex.
J Leukoc Biol, 110, 2021
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK2
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK4
 
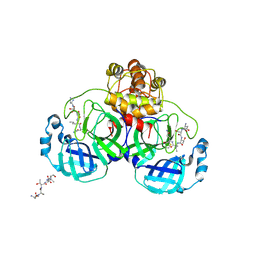 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RZ4
 
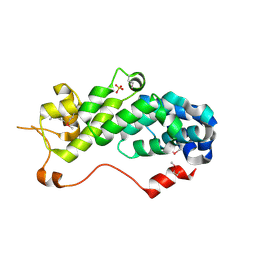 | | Crystal Structure of Human eIF3k | | Descriptor: | Eukaryotic translation initiation factor 3 subunit 11, SULFATE ION | | Authors: | Wei, Z, Zhang, P, Zhou, Z, Gong, W. | | Deposit date: | 2003-12-23 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human eIF3k, the first structure of eIF3 subunits
J.Biol.Chem., 279, 2004
|
|
1TXC
 
 | |
1TW0
 
 | |
1ZXT
 
 | | Crystal Structure of A Viral Chemokine | | Descriptor: | functional macrophage inflammatory protein 1-alpha homolog | | Authors: | Luz, J.G, Yu, M, Su, Y, Wu, Z, Zhou, Z, Sun, R, Wilson, I.A. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of viral macrophage inflammatory protein I encoded by Kaposi's sarcoma-associated herpesvirus at 1.7A.
J.Mol.Biol., 352, 2005
|
|
2LBC
 
 | | solution structure of tandem UBA of USP13 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 13 | | Authors: | Zhang, Y, Zhou, C, Zhou, Z, Song, A, Hu, H. | | Deposit date: | 2011-03-29 | | Release date: | 2012-03-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Domain Analysis Reveals That a Deubiquitinating Enzyme USP13 Performs Non-Activating Catalysis for Lys63-Linked Polyubiquitin.
Plos One, 6, 2011
|
|
2L80
 
 | | Solution Structure of the Zinc Finger Domain of USP13 | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 13, ZINC ION | | Authors: | Zhang, Y, Zhou, C, Zhou, Z, Song, A, Hu, H. | | Deposit date: | 2010-12-28 | | Release date: | 2011-12-14 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Biochemical Characterization of the Ubiquitin Receptors in USP13 Reveals Different Catalytic Activation of Deubiquitination from Its Analogue USP5
To be Published
|
|
1YZC
 
 | |
4LN0
 
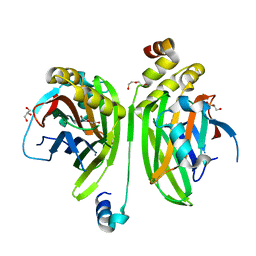 | | Crystal structure of the VGLL4-TEAD4 complex | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Transcription cofactor vestigial-like protein 4, ... | | Authors: | Wang, H, Shi, Z, Zhou, Z. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.896 Å) | | Cite: | A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer.
Cancer Cell, 25, 2014
|
|
2O0F
 
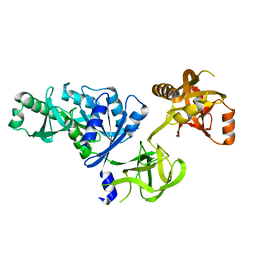 | | Docking of the modified RF3 X-ray structure into cryo-EM map of E.coli 70S ribosome bound with RF3 | | Descriptor: | Peptide chain release factor 3 | | Authors: | Gao, H, Zhou, Z, Rawat, U, Huang, C, Bouakaz, L, Wang, C, Liu, Y, Zavialov, A, Gursky, R, Sanyal, S, Ehrenberg, M, Frank, J, Song, H. | | Deposit date: | 2006-11-27 | | Release date: | 2007-07-24 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (15.5 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
2B9C
 
 | | Structure of tropomyosin's mid-region: bending and binding sites for actin | | Descriptor: | striated-muscle alpha tropomyosin | | Authors: | Brown, J.H, Zhou, Z, Reshetnikova, L, Robinson, H, Yammani, R.D, Tobacman, L.S, Cohen, C. | | Deposit date: | 2005-10-11 | | Release date: | 2006-01-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mid-region of tropomyosin: Bending and binding sites for actin.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
7KOJ
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]-5-{[(prop-2-en-1-yl)carbamoyl]amino}benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder494
to be published
|
|
7JN2
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-03 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2 in complex with PLP_Snyder441
to be published
|
|
7JIV
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder530 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(acryloylamino)-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
7KRX
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441 inhibitor | | Descriptor: | 3-amino-2-methyl-N-[(1R)-1-(naphthalen-1-yl)ethyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | The crystal structure of Papain-Like Protease of SARS CoV-2, C111S mutant, in complex with PLP_Snyder441
to be published
|
|
