6L58
 
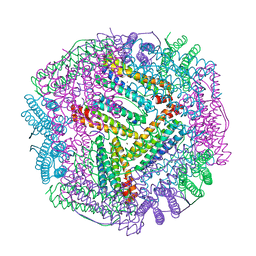 | | Cu(II) loaded Tegillarca granosa M-ferritin soaked with Fe(II) | | Descriptor: | COPPER (II) ION, Ferritin | | Authors: | Jiang, Q.Q, Su, X.R, Ming, T.H, Huan, H.S. | | Deposit date: | 2019-10-22 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.90270972 Å) | | Cite: | Structural Insights Into the Effects of Interactions With Iron and Copper Ions on Ferritin From the Blood Clam Tegillarca granosa.
Front Mol Biosci, 9, 2022
|
|
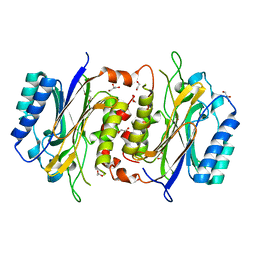
6LED
 
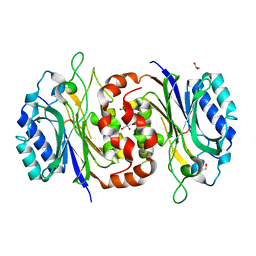 | |
6LE2
 
 | | Structure of D-carbamoylase mutant from Nitratireductor indicus | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-carbamoyl-D-amino-acid hydrolase | | Authors: | Ni, Y, Liu, Y.F, Xu, G.C, Dai, W. | | Deposit date: | 2019-11-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Engineering of D-Carbamoylase Reveals a Key Loop at Substrate Entrance Tunnel
Acs Catalysis, 10, 2020
|
|
8I1V
 
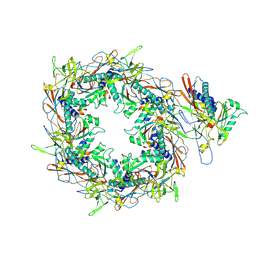 | | The asymmetric unit of P22 procapsid | | Descriptor: | Major capsid protein, Scaffolding protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
 | | The asymmetric unit of P22 empty capsid | | Descriptor: | Major capsid protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|
8HWT
 
 | |
8I99
 
 | | N-carbamoyl-D-amino-acid hydrolase mutant - M4Th3 | | Descriptor: | N-carbamoyl-D-amino-acid hydrolase | | Authors: | Hu, J.M, Ni, Y, Xu, G.C. | | Deposit date: | 2023-02-06 | | Release date: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the Thermostability of a d-Carbamoylase Based on Ancestral Sequence Reconstruction for the Efficient Synthesis of d-Tryptophan.
J.Agric.Food Chem., 71, 2023
|
|
6IS2
 
 | |
6IS3
 
 | |
5HCV
 
 | |
8JAN
 
 | |
8JAJ
 
 | |
6IS1
 
 | |
6IS4
 
 | |
8JD9
 
 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class1 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
8JDA
 
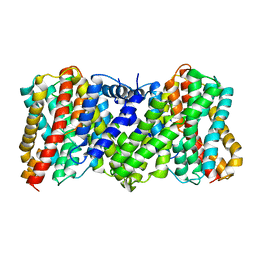 | | Cyro-EM structure of the Na+/H+ antipoter SOS1 from Arabidopsis thaliana,class2 | | Descriptor: | Sodium/hydrogen exchanger 7 | | Authors: | Yang, G.H, Zhang, Y.M, Zhou, J.Q, Jia, Y.T, Xu, X, Fu, P, Wu, H.Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance.
Nat.Plants, 9, 2023
|
|
7E9Q
 
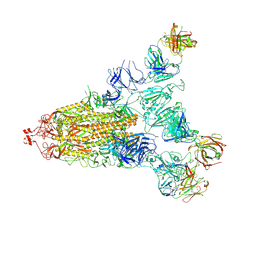 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 out RBD, state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9O
 
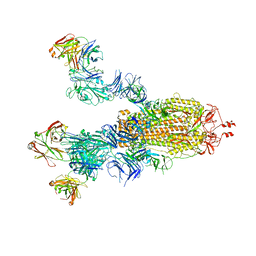 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(3 up RBDs, state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-03-09 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7E9P
 
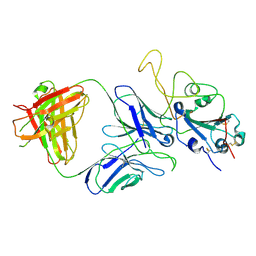 | |
7E9N
 
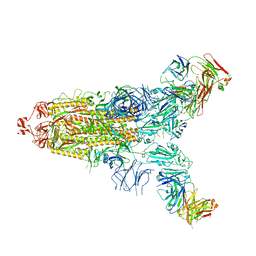 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENG
 
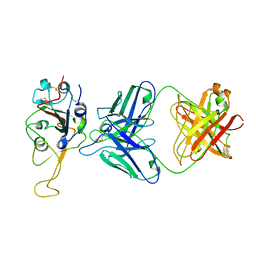 | |
7ENF
 
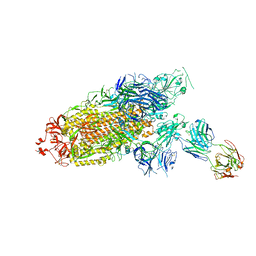 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7F46
 
 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab (state1, local refinement of the RBD, NTD and 35B5 Fab) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-06-17 | | Release date: | 2022-03-23 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (4.79 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
1ZA6
 
 | | The structure of an antitumor CH2-domain-deleted humanized antibody | | Descriptor: | IGG Heavy chain, IGG Light chain | | Authors: | Larson, S.B, Day, J.S, Glaser, S, Braslawsky, G, McPherson, A. | | Deposit date: | 2005-04-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of an Antitumor C(H)2-domain-deleted Humanized Antibody.
J.Mol.Biol., 348, 2005
|
|
