7X52
 
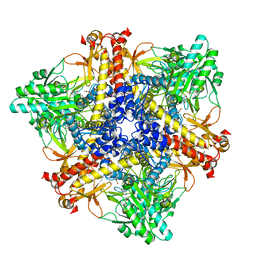 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP complex | | 分子名称: | ACETATE ION, Glutamate decarboxylase, MALONATE ION, ... | | 著者 | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | 登録日 | 2022-03-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4Y
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GABA complex | | 分子名称: | GAMMA-AMINO-BUTANOIC ACID, Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Liu, S, Du, G, Wang, Y, Wen, B, Xin, F. | | 登録日 | 2022-03-03 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X51
 
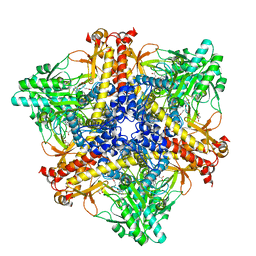 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase BTGAD-PLP-GUA complex | | 分子名称: | GLUTARIC ACID, Glutamate decarboxylase, MALONATE ION, ... | | 著者 | Liu, S, Du, G, Wang, L, Wen, B, Xin, F. | | 登録日 | 2022-03-03 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7X4L
 
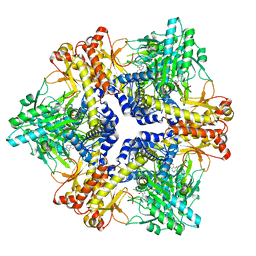 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | 分子名称: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | 登録日 | 2022-03-02 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
7YP0
 
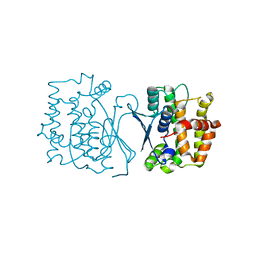 | | Crystal structure of CtGST | | 分子名称: | Glutathione S-transferase | | 著者 | Yang, J, Fan, J.P, Lei, X.G. | | 登録日 | 2022-08-02 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
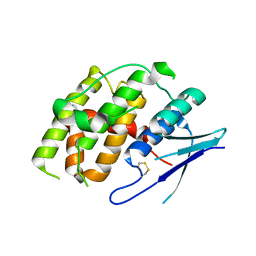 | |
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | 分子名称: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | 著者 | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | 登録日 | 2021-02-21 | | 公開日 | 2021-06-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.59 Å) | | 主引用文献 | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
5WRG
 
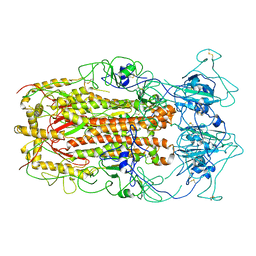 | | SARS-CoV spike glycoprotein | | 分子名称: | Spike glycoprotein | | 著者 | Gui, M, Song, W, Xiang, Y, Wang, X. | | 登録日 | 2016-12-01 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding.
Cell Res., 27, 2017
|
|
5XLR
 
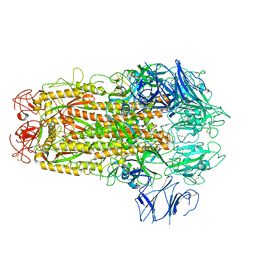 | |
5XRR
 
 | | Crystal structure of FUS (54-59) SYSSYG | | 分子名称: | RNA-binding protein FUS, ZINC ION | | 著者 | Zhao, M, Gui, X, Li, D, Liu, C. | | 登録日 | 2017-06-09 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5Y7L
 
 | |
8IJS
 
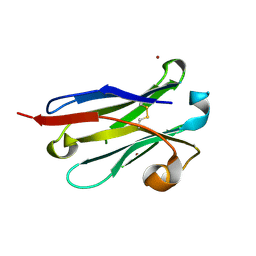 | | anti-VEGF nanobody mutant | | 分子名称: | ZINC ION, anti-VEGF nanobody | | 著者 | Qian, F, Zhu, S.Q. | | 登録日 | 2023-02-28 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IJZ
 
 | | anti-VEGF mutant | | 分子名称: | anti-VEGF nanobody | | 著者 | Qian, F, Zhu, S.Q. | | 登録日 | 2023-02-28 | | 公開日 | 2023-12-27 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | 主引用文献 | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
8IIU
 
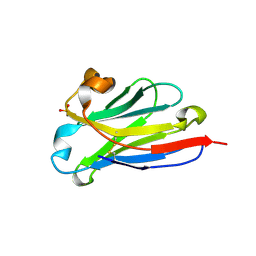 | | anti-VEGF nanobody | | 分子名称: | SULFATE ION, anti-VEGF nanobody | | 著者 | Qian, F, Zhu, S.Q. | | 登録日 | 2023-02-24 | | 公開日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Polymorphic nanobody crystals as long-acting intravitreal therapy for wet age-related macular degeneration.
Bioeng Transl Med, 8, 2023
|
|
5ZGD
 
 | | hnRNPA1 reversible amyloid core GFGGNDNFG (residues 209-217) determined by X-ray | | 分子名称: | GLY-PHE-GLY-GLY-ASN-ASP-ASN-PHE-GLY | | 著者 | Gui, X, Xie, M, Zhao, M, Luo, F, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-08 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.401 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZGL
 
 | | hnRNP A1 segment GGGYGGS (residues 234-240) | | 分子名称: | 7-mer peptide from Heterogeneous nuclear ribonucleoprotein A1 | | 著者 | Xie, M, Luo, F, Gui, X, Zhao, M, He, J, Li, D, Liu, C. | | 登録日 | 2018-03-09 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly.
Nat Commun, 10, 2019
|
|
5ZMA
 
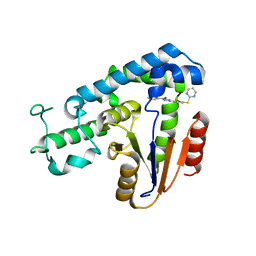 | |
6AAY
 
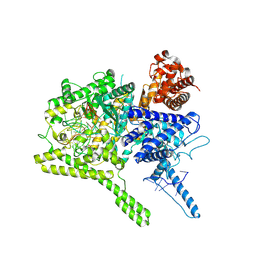 | | the Cas13b binary complex | | 分子名称: | Bergeyella zoohelcum Cas13b (R1177A) mutant, RNA (52-MER) | | 著者 | Bo, Z, Weiwei, Y, Yangmiao, Y, Ouyang, S.Y. | | 登録日 | 2018-07-19 | | 公開日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural insights into Cas13b-guided CRISPR RNA maturation and recognition.
Cell Res., 28, 2018
|
|
7DQ9
 
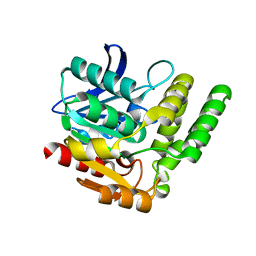 | |
7F26
 
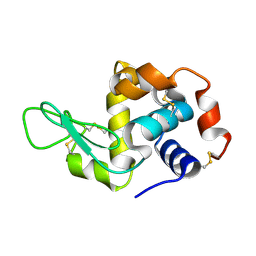 | | Crystal structure of lysozyme | | 分子名称: | Lysozyme C | | 著者 | Liang, M. | | 登録日 | 2021-06-10 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Novel combined crystallization plate for high-throughput crystal screening and in situ data collection at a crystallography beamline.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7F3N
 
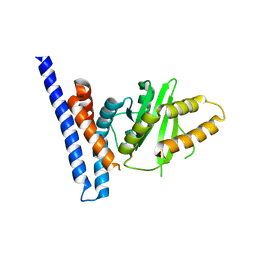 | | Structure of PopP2 in apo form | | 分子名称: | Type III effector protein popp2 | | 著者 | Xia, Y, Zhang, Z.M. | | 登録日 | 2021-06-16 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.351856 Å) | | 主引用文献 | Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase.
Nat Commun, 12, 2021
|
|
7F1E
 
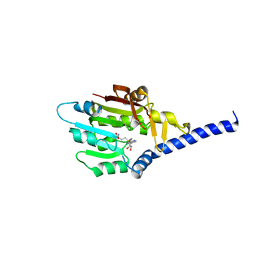 | | Structure of METTL6 bound with SAM | | 分子名称: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | 著者 | Li, S, Liao, S, Xu, C. | | 登録日 | 2021-06-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.589 Å) | | 主引用文献 | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7FH7
 
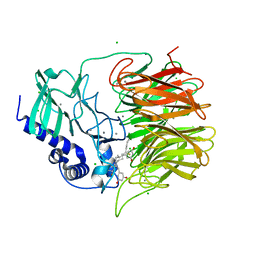 | | Friedel-Crafts alkylation enzyme CylK mutant Y37F | | 分子名称: | 5-[(2S,7R)-7-fluoranyl-2-methyl-undecyl]benzene-1,3-diol, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wang, H.Q, Wei, Z, Xiang, Z. | | 登録日 | 2021-07-29 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
7FH6
 
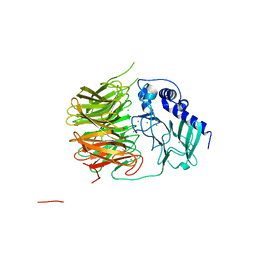 | | Friedel-Crafts alkylation enzyme CylK | | 分子名称: | CALCIUM ION, CHLORIDE ION, CylK, ... | | 著者 | Liu, L, Wang, H.Q, Xiang, Z. | | 登録日 | 2021-07-29 | | 公開日 | 2022-02-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
7FH8
 
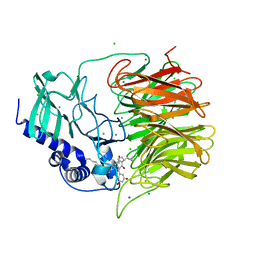 | | Friedel-Crafts alkylation enzyme CylK mutant H391A | | 分子名称: | 2-[(5S,10S)-11-[3,5-bis(oxidanyl)phenyl]-10-methyl-undecan-5-yl]-5-[(2S,7R)-7-fluoranyl-2-methyl-undecyl]benzene-1,3-diol, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Wang, H.Q, Wei, Z, Xiang, Z. | | 登録日 | 2021-07-29 | | 公開日 | 2022-02-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural Basis for the Friedel-Crafts Alkylation in Cylindrocyclophane Biosynthesis
ACS Catal., 12, 2022
|
|
