5CDI
 
 | | Chloroplast chaperonin 60b1 of Chlamydomonas | | Descriptor: | Chaperonin 60B1 | | Authors: | Zhang, S, Zhou, H, Yu, F, Gao, F, He, J, Liu, C. | | Deposit date: | 2015-07-04 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.807 Å) | | Cite: | Structural insight into the cooperation of chloroplast chaperonin subunits
Bmc Biol., 14, 2016
|
|
5CMA
 
 | | Anti-B7-H3 monoclonal antibody ch8H9 Fab fragment | | Descriptor: | Antibody ch8H9 Fab heavy chain, Antibody ch8H9 Fab light chain | | Authors: | Ahmed, M, Goldgur, Y, Cheng, M, Cheung, N.K. | | Deposit date: | 2015-07-16 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Humanized Affinity-matured Monoclonal Antibody 8H9 Has Potent Antitumor Activity and Binds to FG Loop of Tumor Antigen B7-H3.
J.Biol.Chem., 290, 2015
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
2Q8T
 
 | | Crystal Structure of the CC chemokine CCL14 | | Descriptor: | CCL14 | | Authors: | Blain, K.Y. | | Deposit date: | 2007-06-11 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Functional Characterization of CC Chemokine CCL14
Biochemistry, 46, 2007
|
|
2Q6F
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease in complex with a Michael acceptor inhibitor N3 | | Descriptor: | Infectious bronchitis virus (IBV) main protease, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q8R
 
 | |
2Q6D
 
 | | Crystal structure of infectious bronchitis virus (IBV) main protease | | Descriptor: | Infectious bronchitis virus (IBV) main protease | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-04 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
2Q6G
 
 | | Crystal structure of SARS-CoV main protease H41A mutant in complex with an N-terminal substrate | | Descriptor: | Polypeptide chain, severe acute respiratory syndrome coronavirus (SARS-CoV) | | Authors: | Xue, X.Y, Yang, H.T, Xue, F, Bartlam, M, Rao, Z.H. | | Deposit date: | 2007-06-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of two coronavirus main proteases: implications for substrate binding and antiviral drug design.
J.Virol., 82, 2008
|
|
8HQ7
 
 | |
8HPF
 
 | |
8HPU
 
 | |
8HPQ
 
 | |
8HP9
 
 | |
8HPV
 
 | |
4FJQ
 
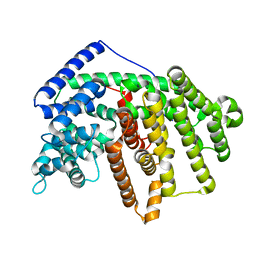 | | Crystal Structure of an alpha-Bisabolol synthase | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Jianxu, L, Peng, Z. | | Deposit date: | 2012-06-12 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0001 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
4GAX
 
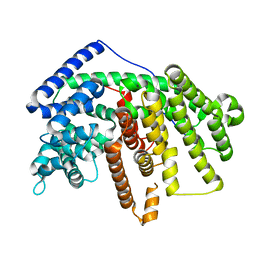 | | Crystal Structure of an alpha-Bisabolol synthase mutant | | Descriptor: | Amorpha-4,11-diene synthase | | Authors: | Li, J, Peng, Z. | | Deposit date: | 2012-07-26 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9948 Å) | | Cite: | Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency.
Biochem.J., 451, 2013
|
|
8IXB
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXG
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXD
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXE
 
 | | GMPCPP-Alpha1C/Beta2A-microtubule decorated with kinesin seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXA
 
 | | GMPCPP-Alpha1A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8IXF
 
 | | GMPCPP-Alpha4A/Beta2A-microtubule decorated with kinesin non-seam region | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-1 heavy chain, ... | | Authors: | Zheng, W, Zhao, Q.Y, Diao, L, Bao, L, Cong, Y. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-16 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8ILF
 
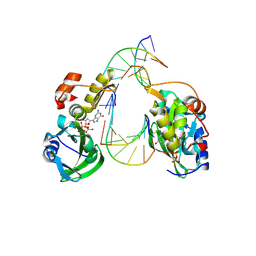 | | The crystal structure of dGTPalphaSe-Sp:DNApre-II:Pol X substrate ternary complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*GP*AP*TP*CP*CP*A)-3'), MANGANESE (II) ION, Repair DNA polymerase X, ... | | Authors: | Qin, T, Zhao, Q.W, Gan, J.H, Huang, Z. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Polymerase Mechanism via a Chiral Center Generated with a Single Selenium Atom.
Int J Mol Sci, 24, 2023
|
|
7Y04
 
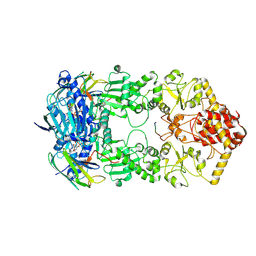 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
2AF9
 
 | | Crystal Structure analysis of GM2-Activator protein complexed with phosphatidylcholine | | Descriptor: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, LAURIC ACID, ... | | Authors: | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
