8UQ8
 
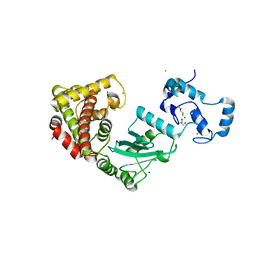 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | 分子名称: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
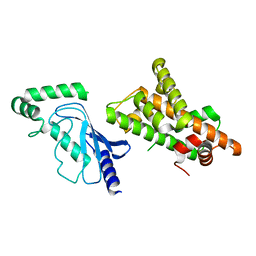 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.893 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
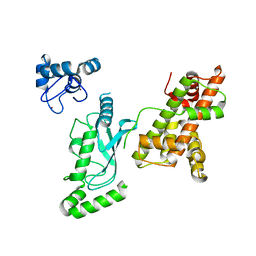 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5H3D
 
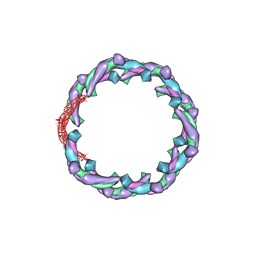 | | Helical structure of membrane tubules decorated by ACAP1 (BARPH doamin) protein by cryo-electron microscopy and MD simulation | | 分子名称: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | 著者 | Chan, C, Pang, X.Y, Zhang, Y, Sun, F, Fan, J. | | 登録日 | 2016-10-22 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (14 Å) | | 主引用文献 | ACAP1 assembles into an unusual protein lattice for membrane deformation through multiple stages.
Plos Comput.Biol., 15, 2019
|
|
6OAI
 
 | | Crystal structure of P[6] rotavirus vp8* complexed with LNFPI | | 分子名称: | Protease-sensitive outer capsid protein, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Xu, S, Liu, Y, Jiang, X, Kennedy, M.A. | | 登録日 | 2019-03-16 | | 公開日 | 2020-03-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens.
Plos Pathog., 16, 2020
|
|
5FB1
 
 | |
5FB0
 
 | |
3HD3
 
 | | High resolution crystal structure of cruzain bound to the vinyl sulfone inhibitor SMDC-256047 | | 分子名称: | (1R,2R)-2-[(4-chlorophenyl)carbonyl]-N-{(1S)-1-[2-(phenylsulfonyl)ethyl]pentyl}cyclohexanecarboxamide, 1,2-ETHANEDIOL, Cruzipain, ... | | 著者 | Kerr, I.D, Debnath, M, Brinen, L.S. | | 登録日 | 2009-05-06 | | 公開日 | 2009-10-06 | | 最終更新日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Novel non-peptidic vinylsulfones targeting the S2 and S3 subsites of parasite cysteine proteases.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5XVA
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH216 | | 分子名称: | ETHANOL, Serine/threonine-protein kinase PAK 4, [6-chloranyl-4-[(5-methyl-1H-pyrazol-3-yl)amino]quinazolin-2-yl]-[(3R)-3-methylpiperazin-1-yl]methanone | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVG
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH226 | | 分子名称: | 1,2-ETHANEDIOL, ETHANOL, Serine/threonine-protein kinase PAK 4, ... | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | 分子名称: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | 著者 | Zhao, F, Li, H. | | 登録日 | 2017-06-27 | | 公開日 | 2018-02-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
6L2T
 
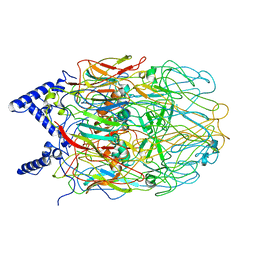 | |
6LT8
 
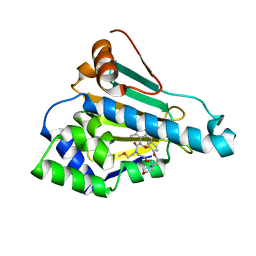 | | HSP90 in complex with KW-2478 | | 分子名称: | 2-[2-ethyl-6-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]carbonyl-3,5-bis(oxidanyl)phenyl]-~{N},~{N}-bis(2-methoxyethyl)ethanamide, Heat shock protein HSP 90-alpha | | 著者 | Cao, H.L. | | 登録日 | 2020-01-21 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.593 Å) | | 主引用文献 | Anti-NSCLC activity in vitro of Hsp90 N inhibitor KW-2478 and complex crystal structure determination of Hsp90 N -KW-2478.
J.Struct.Biol., 213, 2021
|
|
6LR9
 
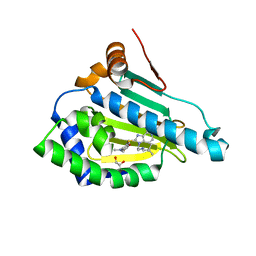 | | HSP90 in complex with Debio0932 | | 分子名称: | 2-[[6-(dimethylamino)-1,3-benzodioxol-5-yl]sulfanyl]-1-[2-(2,2-dimethylpropylamino)ethyl]imidazo[4,5-c]pyridin-4-amine, GLYCEROL, Heat shock protein HSP 90-alpha | | 著者 | Cao, H.L. | | 登録日 | 2020-01-15 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.196 Å) | | 主引用文献 | Complex crystal structure determination and anti-non-small-cell lung cancer activity of the Hsp90 N inhibitor Debio0932.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6D0L
 
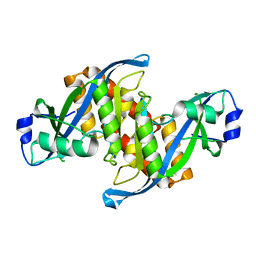 | | Structure of human TIRR | | 分子名称: | Tudor-interacting repair regulator protein | | 著者 | Cui, G, Botuyan, M.V, Mer, G. | | 登録日 | 2018-04-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CO1
 
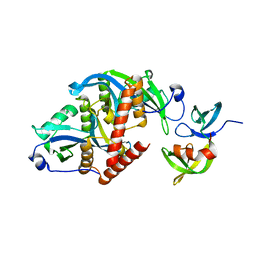 | |
1HP4
 
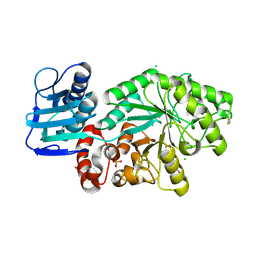 | |
6CO2
 
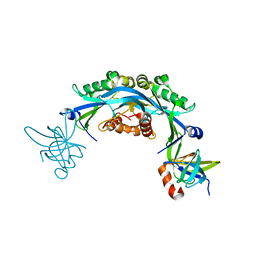 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | 分子名称: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | 著者 | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | 登録日 | 2018-03-10 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7XIY
 
 | | SARS-CoV-2 Omicron BA.3 variant spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Wang, L. | | 登録日 | 2022-04-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIX
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Wang, L. | | 登録日 | 2022-04-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Wang, L. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Wang, L. | | 登録日 | 2022-04-14 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XIZ
 
 | |
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Wang, X, Wang, L. | | 登録日 | 2022-04-29 | | 公開日 | 2022-07-13 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7X6A
 
 | |
