2VH4
 
 | | Structure of a loop C-sheet serpin polymer | | 分子名称: | TENGPIN | | 著者 | Zhang, Q, Law, R.H.P, Bottomley, S.P, Whisstock, J.C, Buckle, A.M. | | 登録日 | 2007-11-19 | | 公開日 | 2008-01-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A Structural Basis for Loop C-Sheet Polymerization in Serpins.
J.Mol.Biol., 376, 2008
|
|
1I9F
 
 | | STRUCTURAL CHARACTERIZATION OF THE COMPLEX OF THE REV RESPONSE ELEMENT RNA WITH A SELECTED PEPTIDE | | 分子名称: | REV RESPONSE ELEMENT RNA, RSG-1.2 PEPTIDE | | 著者 | Zhang, Q, Harada, K, Cho, H.S, Frankel, A, Wemmer, D.E. | | 登録日 | 2001-03-19 | | 公開日 | 2001-05-25 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural characterization of the complex of the Rev response element RNA with a selected peptide.
Chem.Biol., 8, 2001
|
|
2DO8
 
 | | Solution Structure of UPF0301 protein HD_1794 | | 分子名称: | UPF0301 protein HD_1794 | | 著者 | Zhang, Q, Liu, G, Shastry, R, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T.R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-04-27 | | 公開日 | 2006-05-25 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of UPF0301 protein HD_1794
To be published
|
|
6V3X
 
 | |
6V3Y
 
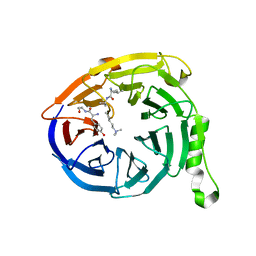 | |
1PQQ
 
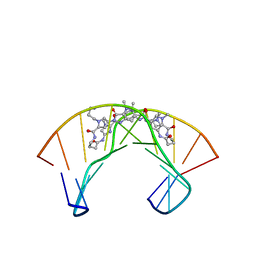 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | 分子名称: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | 著者 | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | 登録日 | 2003-06-18 | | 公開日 | 2004-06-29 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | 分子名称: | Hemocyanin isoform 1 | | 著者 | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | 登録日 | 2013-02-20 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
2H3J
 
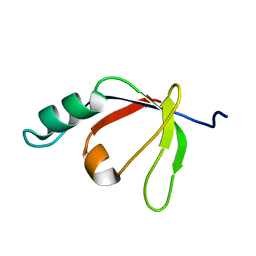 | | Solution NMR Structure of Protein PA4359 from Pseudomonas aeruginosa: Northeast Structural Genomics Consortium Target PaT89 | | 分子名称: | Hypothetical protein PA4359 | | 著者 | Zhang, Q, Liu, G, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-05-22 | | 公開日 | 2006-06-20 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Hypothetical protein PA4359: Northest Structural Genomics Target PaT89
To be Published
|
|
2L3E
 
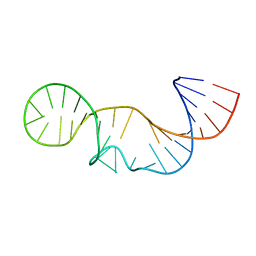 | | Solution structure of P2a-J2a/b-P2b of human telomerase RNA | | 分子名称: | 35-MER | | 著者 | Zhang, Q, Kim, N, Peterson, R.D, Wang, Z, Feigon, J. | | 登録日 | 2010-09-13 | | 公開日 | 2010-11-17 | | 最終更新日 | 2011-07-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inaugural Article: Structurally conserved five nucleotide bulge determines the overall topology of the core domain of human telomerase RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | 分子名称: | Putative periplasmic protein | | 著者 | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2006-10-25 | | 公開日 | 2006-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
5DV2
 
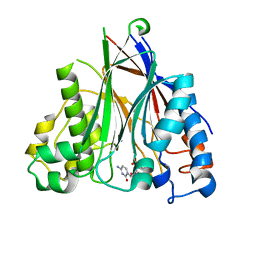 | |
5DV4
 
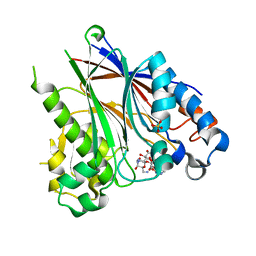 | |
8W59
 
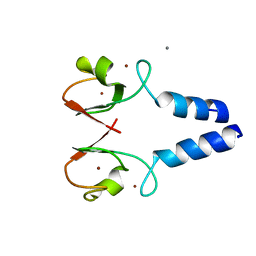 | |
8W5A
 
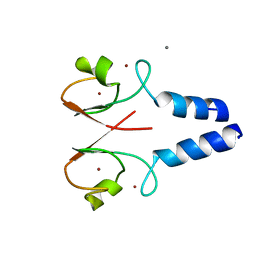 | |
4FWJ
 
 | |
4FWE
 
 | |
4FWF
 
 | | Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.1, Lysine-specific histone demethylase 1B, ... | | 著者 | Zhang, Q, Chen, Z. | | 登録日 | 2012-07-01 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
8GV9
 
 | | The cryo-EM structure of hAE2 with chloride ion | | 分子名称: | Anion exchange protein 2, CHLORIDE ION | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GV8
 
 | | The cryo-EM structure of hAE2 with DIDS | | 分子名称: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Anion exchange protein 2 | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVE
 
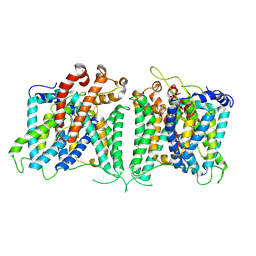 | | The asymmetry structure of hAE2 | | 分子名称: | Anion exchange protein 2 | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-15 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVF
 
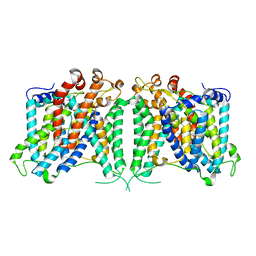 | | The outward-facing structure of hAE2 in basic pH | | 分子名称: | Anion exchange protein 2 | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-15 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVC
 
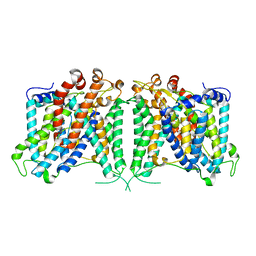 | | The cryo-EM structure of hAE2 with bicarbonate | | 分子名称: | Anion exchange protein 2, BICARBONATE ION | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVH
 
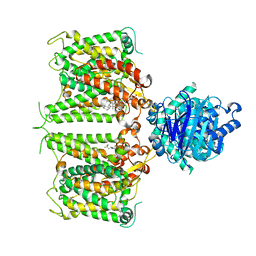 | | Human AE2 in acidic KNO3 | | 分子名称: | Anion exchange protein 2, CHOLESTEROL HEMISUCCINATE | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-15 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
8GVA
 
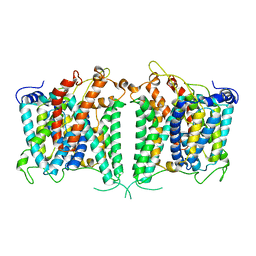 | | The intermediate structure of hAE2 in basic pH | | 分子名称: | Anion exchange protein 2 | | 著者 | Zhang, Q, Jian, L, Yao, D, Rao, B, Hu, K, Xia, Y, Cao, Y. | | 登録日 | 2022-09-14 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | The structural basis of the pH-homeostasis mediated by the Cl - /HCO 3 - exchanger, AE2.
Nat Commun, 14, 2023
|
|
7F40
 
 | | Lysophospholipid acyltransferase LPCAT3 in a complex with Arachidonoyl-CoA | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, LPCAT3, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenethioate | | 著者 | Zhang, Q, Yao, D, Rao, B, Li, S, Jian, L, Chen, Y, Hu, K, Xia, Y, Shen, Y, Cao, Y. | | 登録日 | 2021-06-17 | | 公開日 | 2021-12-01 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.49 Å) | | 主引用文献 | The structural basis for the phospholipid remodeling by lysophosphatidylcholine acyltransferase 3.
Nat Commun, 12, 2021
|
|
