4KYK
 
 | | Crystal structure of mouse glyoxalase I complexed with indomethacin | | 分子名称: | INDOMETHACIN, Lactoylglutathione lyase, ZINC ION | | 著者 | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | 登録日 | 2013-05-29 | | 公開日 | 2013-08-07 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
4KYH
 
 | | Crystal structure of mouse glyoxalase I complexed with zopolrestat | | 分子名称: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Lactoylglutathione lyase, ZINC ION | | 著者 | Zhai, J, Yuan, M, Zhang, L, Chen, Y, Zhang, H, Chen, S, Zhao, Y. | | 登録日 | 2013-05-29 | | 公開日 | 2013-08-07 | | 最終更新日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Zopolrestat as a human glyoxalase I inhibitor and its structural basis.
Chemmedchem, 8, 2013
|
|
4OPN
 
 | |
4RVP
 
 | | Crystal structure of superoxide dismutase from sedum alfredii | | 分子名称: | ZINC ION, superoxide dismutase | | 著者 | Qiu, R, Li, C, Zhai, J, Tang, L, Zhang, H, Yuan, M, Hu, X. | | 登録日 | 2014-11-27 | | 公開日 | 2015-12-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The positive effects of Cd and Cd-Zn relationship in the
Zn-related physiological processes involved in growth in the Zn/Cd hyperaccumulator Sedum alfredii
To be Published
|
|
4X2A
 
 | | Crystal structure of mouse glyoxalase I complexed with baicalein | | 分子名称: | 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one, Lactoylglutathione lyase, ZINC ION | | 著者 | Zhang, H, Zhai, J, Zhang, L, Li, C, Zhao, Y, Hu, X. | | 登録日 | 2014-11-26 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | In Vitro Inhibition of Glyoxalase І by Flavonoids: New Insights from Crystallographic Analysis.
Curr Top Med Chem, 16, 2016
|
|
4PV5
 
 | | Crystal structure of mouse glyoxalase I in complexed with 18-beta-glycyrrhetinic acid | | 分子名称: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, Lactoylglutathione lyase, ZINC ION | | 著者 | Zhang, H, Zhai, J, Zhang, L.P, Zhao, Y.N, Li, C, Hu, X.P. | | 登録日 | 2014-03-15 | | 公開日 | 2015-03-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for 18-beta-glycyrrhetinic acid as a novel non-GSH analog glyoxalase I inhibitor
Acta Pharmacol.Sin., 36, 2015
|
|
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | 分子名称: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | 登録日 | 2012-08-23 | | 公開日 | 2013-08-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
4I5X
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Flufenamic acid | | 分子名称: | 2-[[3-(TRIFLUOROMETHYL)PHENYL]AMINO] BENZOIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhang, L, Zheng, X, Chen, S, Zhai, J, Zhang, H, Zhao, Y. | | 登録日 | 2012-11-29 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
4JIH
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Epalrestat | | 分子名称: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-[(2E)-2-methyl-3-phenylprop-2-en-1-ylidene]-4-oxo-2-thioxo-1,3-thiazolidin-3-yl}acetic acid | | 著者 | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | 登録日 | 2013-03-06 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIR
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Epalrestat | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | 著者 | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X. | | 登録日 | 2013-03-06 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JII
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | 分子名称: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | 登録日 | 2013-03-06 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
7RRB
 
 | | IDO1 IN COMPLEX WITH COMPOUND 9 | | 分子名称: | 3-[4-(6-cyclopropylpyridin-3-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | 著者 | Lesburg, C.A. | | 登録日 | 2021-08-09 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRC
 
 | | IDO1 IN COMPLEX WITH COMPOUND 14 | | 分子名称: | Indoleamine 2,3-dioxygenase 1, N-(4-fluorophenyl)-3-{4-[4-(hydroxymethyl)-6-(trifluoromethyl)pyridin-3-yl]phenyl}oxetane-3-carboxamide | | 著者 | Lesburg, C.A. | | 登録日 | 2021-08-09 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-05-11 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Oxetane Promise Delivered: Discovery of Long-Acting IDO1 Inhibitors Suitable for Q3W Oral or Parenteral Dosing.
J.Med.Chem., 65, 2022
|
|
7RRD
 
 | | IDO1 IN COMPLEX WITH COMPOUND S-1 | | 分子名称: | 3-[4-(1H-benzimidazol-2-yl)phenyl]-N-(4-fluorophenyl)oxetane-3-carboxamide, Indoleamine 2,3-dioxygenase 1 | | 著者 | Lesburg, C.A. | | 登録日 | 2021-08-09 | | 公開日 | 2022-08-24 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Oxetane promise delivered: discovery of long acting IDO1 inhibitors suitable for Q3W oral or parenteral dosing
To Be Published
|
|
7L4C
 
 | | Crystal structure of the DRM2-CTT DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*AP*TP*TP*AP*AP*TP*(C49)P*TP*TP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*TP*AP*AP*TP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-18 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4K
 
 | | Crystal structure of the DRM2-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4N
 
 | | Crystal structure of the DRM2 (C397R)-CCG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*CP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*GP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.247 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4M
 
 | | Crystal structure of the DRM2-CCT DNA complex | | 分子名称: | DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), DNA (5'-D(P*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*CP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4F
 
 | | Crystal structure of the DRM2-CAT DNA complex | | 分子名称: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*AP*TP*TP*CP*CP*TP*CP*CP*TP*(C49)P*AP*TP*CP*CP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*GP*GP*AP*TP*GP*AP*GP*GP*AP*GP*GP*AP*AP*T)-3'), ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
7L4H
 
 | | Crystal structure of the DRM2-CTG DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*TP*CP*CP*TP*AP*AP*TP*(C49)P*TP*GP*AP*AP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*TP*TP*CP*AP*GP*AP*TP*TP*AP*GP*GP*AP*AP*T)-3'), DNA (cytosine-5)-methyltransferase DRM2, ... | | 著者 | Fang, J, Song, J. | | 登録日 | 2020-12-19 | | 公開日 | 2021-08-04 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Substrate deformation regulates DRM2-mediated DNA methylation in plants.
Sci Adv, 7, 2021
|
|
4YVV
 
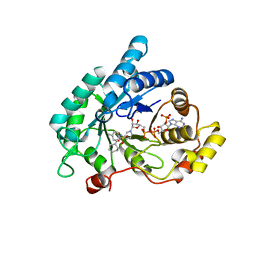 | | Crystal structure of AKR1C3 complexed with glibenclamide | | 分子名称: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVX
 
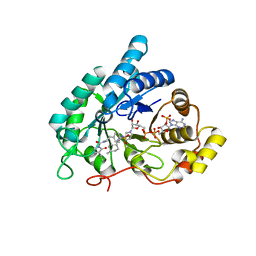 | | Crystal structure of AKR1C3 complexed with glimepiride | | 分子名称: | 3-ethyl-4-methyl-N-[2-(4-{[(cis-4-methylcyclohexyl)carbamoyl]sulfamoyl}phenyl)ethyl]-2-oxo-2,5-dihydro-1H-pyrrole-1-car boxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4YVP
 
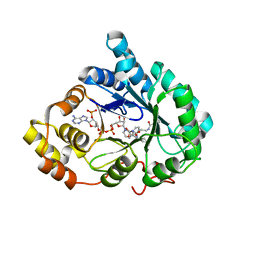 | | Crystal Structure of AKR1C1 complexed with glibenclamide | | 分子名称: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zheng, X, Zhang, H, Hu, X. | | 登録日 | 2015-03-20 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
4ZFC
 
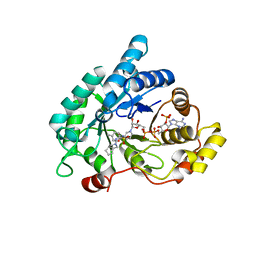 | | Crystal structure of AKR1C3 complexed with glicazide | | 分子名称: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zhao, Y, Zhrng, X, Hu, X. | | 登録日 | 2015-04-21 | | 公開日 | 2015-11-25 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
3RX4
 
 | | Crystal Structure of Human Aldose Reductase complexed with Sulindac Sulfide | | 分子名称: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfanylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Chen, J, Luo, H, Hu, X. | | 登録日 | 2011-05-10 | | 公開日 | 2011-11-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
