6L8F
 
 | |
6L8E
 
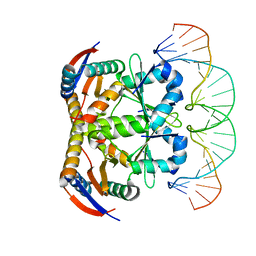 | | Crystal structure of heterohexameric YoeB-YefM complex bound to 26bp-DNA | | Descriptor: | DNA (26-mer), YefM Antitoxin, YoeB toxin | | Authors: | Yue, J, Xue, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distinct oligomeric structures of the YoeB-YefM complex provide insights into the conditional cooperativity of type II toxin-antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
6L8G
 
 | |
7CUA
 
 | | The structure of YoeB dimer from Staphylococcus aureus | | Descriptor: | SULFATE ION, YoeB | | Authors: | Yue, J, Xue, L. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct oligomeric structures of the YoeB-YefM complex provide insights into the conditional cooperativity of type II toxin-antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
7V5Y
 
 | |
7V5Z
 
 | |
7V6W
 
 | | Crystal structure of heterohexameric Sa2YoeB-Sa2YefM complex bound to 26bp-DNA | | Descriptor: | Antitoxin, DNA (25-MER), DNA (26-MER), ... | | Authors: | Xue, L, Khan, M.H, Yue, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The two paralogous copies of the YoeB-YefM toxin-antitoxin module in Staphylococcus aureus differ in DNA binding and recognition patterns.
J.Biol.Chem., 298, 2022
|
|
7W02
 
 | | Cryo-EM structure of ATP-bound ABCA3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7W01
 
 | | Cryo-EM structure of nucleotide-free ABCA3 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xie, T, Zhang, Z.K, Yue, J, Gong, X. | | Deposit date: | 2021-11-17 | | Release date: | 2022-04-27 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of the human surfactant lipid transporter ABCA3.
Sci Adv, 8, 2022
|
|
7D8F
 
 | | The crystal structure of ScNTM1 in complex with SAH | | Descriptor: | Alpha N-terminal protein methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Zhang, H.Y, Yue, J, Zhu, Z.L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural Basis for Peptide Binding of Alpha-N Terminal Methyltransferase from Saccharomyces cerevisiae
Crystallography Reports, 66, 2021
|
|
7D8D
 
 | |
4Z6G
 
 | | Structure of NT domain | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, PHOSPHATE ION | | Authors: | Yang, F, Zhang, Y. | | Deposit date: | 2015-04-05 | | Release date: | 2016-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | In vivo epidermal migration requires focal adhesion targeting of ACF7.
Nat Commun, 7, 2016
|
|
5X57
 
 | | Structure of GAR domain of ACF7 | | Descriptor: | Microtubule-actin cross-linking factor 1, isoforms 1/2/3/5, NICKEL (II) ION | | Authors: | Yang, F, Wang, T, Zhang, Y, Wu, X.Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | ACF7 regulates inflammatory colitis and intestinal wound response by orchestrating tight junction dynamics.
Nat Commun, 8, 2017
|
|
2EW6
 
 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | (2E)-3-(3,4-DIHYDROXYPHENYL)-N-[2-(4-HYDROXYPHENYL)ETHYL]ACRYLAMIDE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW7
 
 | | Crystal Structure of Helicobacter Pylori peptide deformylase | | Descriptor: | COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
2EW5
 
 | | Structure of Helicobacter Pylori peptide deformylase in complex with inhibitor | | Descriptor: | 4-{(1E)-3-OXO-3-[(2-PHENYLETHYL)AMINO]PROP-1-EN-1-YL}-1,2-PHENYLENE DIACETATE, COBALT (II) ION, peptide deformylase | | Authors: | Cai, J. | | Deposit date: | 2005-11-02 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: reverse docking, enzymatic assay, and X-ray crystallography validation
Protein Sci., 15, 2006
|
|
7BPB
 
 | | Human AAA+ ATPase VCP mutant - T76E, AMP-PNP bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-22 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP8
 
 | | Human AAA+ ATPase VCP mutant - T76A, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BP9
 
 | | Human AAA+ ATPase VCP mutant - T76E, ADP-bound form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7BPA
 
 | | Human AAA+ ATPase VCP mutant - T76A, AMP-PNP-bound form, Conformation I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Yang, C, Zhang, H. | | Deposit date: | 2020-03-21 | | Release date: | 2021-03-31 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The phosphorylation and dephosphorylation switch of VCP/p97 regulates the architecture of centrosome and spindle.
Cell Death Differ., 2022
|
|
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6AIM
 
 | | Cab2 mutant H337A complex with phosphopantothenate-cysteine | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl-L-cysteine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AI9
 
 | | Cab2 mutant-H337A complex with phosphopantothenate | | Descriptor: | N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanine, Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AI8
 
 | | Cab2 mutant-H337A | | Descriptor: | GLYCEROL, Phosphopantothenate--cysteine ligase CAB2, SULFATE ION | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-22 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
6AIP
 
 | | Cab2 mutant-H337A complex with phosphopantothenoylcystine | | Descriptor: | (5R,12R,17R)-17-amino-12-carboxy-1,1,5-trihydroxy-4,4-dimethyl-6,10-dioxo-2-oxa-14,15-dithia-7,11-diaza-1-phosphaoctadecan-18-oic acid 1-oxide (non-preferred name), Phosphopantothenate--cysteine ligase CAB2 | | Authors: | Zheng, P, Zhu, Z. | | Deposit date: | 2018-08-24 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystallographic Analysis of the Catalytic Mechanism of Phosphopantothenoylcysteine Synthetase from Saccharomyces cerevisiae.
J. Mol. Biol., 431, 2019
|
|
