2HFN
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | FLAVIN MONONUCLEOTIDE, Synechocystis Photoreceptor (Slr1694) | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
2HFO
 
 | | Crystal Structures of the Synechocystis Photoreceptor Slr1694 Reveal Distinct Structural States Related to Signaling | | Descriptor: | Activator of photopigment and puc expression, FLAVIN MONONUCLEOTIDE | | Authors: | Yuan, H, Anderson, S, Masuda, S, Dragnea, V, Moffat, K, Bauer, C.E. | | Deposit date: | 2006-06-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Synechocystis photoreceptor Slr1694 reveal distinct structural states related to signaling.
Biochemistry, 45, 2006
|
|
5GOL
 
 | |
5H6W
 
 | |
1D56
 
 | |
1D57
 
 | |
3MZI
 
 | |
3TOB
 
 | |
3TO9
 
 | | Crystal structure of yeast Esa1 E338Q HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | 1,2-ETHANEDIOL, CACODYLIC ACID, COENZYME A, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TOA
 
 | | Human MOF crystal structure with active site lysine partially acetylated | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
3TO6
 
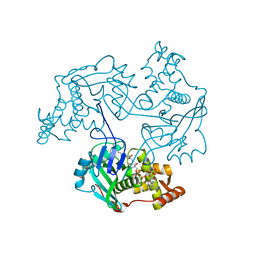 | |
3TO7
 
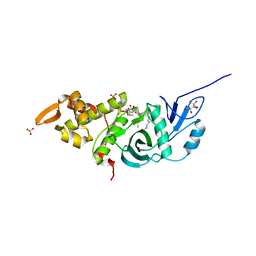 | | Crystal structure of yeast Esa1 HAT domain bound to coenzyme A with active site lysine acetylated | | Descriptor: | CACODYLIC ACID, COENZYME A, GLYCEROL, ... | | Authors: | Yuan, H, Ding, E.C, Marmorstein, R. | | Deposit date: | 2011-09-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MYST protein acetyltransferase activity requires active site lysine autoacetylation.
Embo J., 31, 2011
|
|
9JMI
 
 | |
9JMO
 
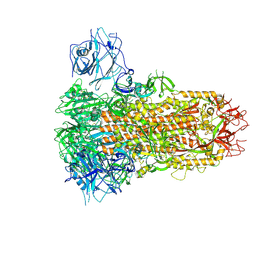 | | Cryo-EM structure of Japan-BatCoV (Vs-CoV-1) S-trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yuan, H, Xiong, X. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
9JMM
 
 | | Cryo-EM structure of the SE-PangolinCoV (MjHKU4r-CoV-1) RBD in complex with human DPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form, ... | | Authors: | Yuan, H, Xiong, X. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
9JMF
 
 | | Cryo-EM structure of SA-BatCoV (Neoromicia/PML-PHE1/RSA/2011) S-trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yuan, H, Xiong, X. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
9JMJ
 
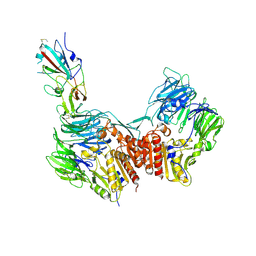 | | Cryo-EM structure of the GD-BatCoV (BtCoV/Ii/GD/2014-422) RBD in complex with human DPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 soluble form,Isoform 1 of Immunoglobulin heavy constant gamma 1, ... | | Authors: | Yuan, H, Xiong, X. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
9JMG
 
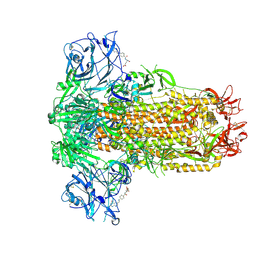 | |
9JMN
 
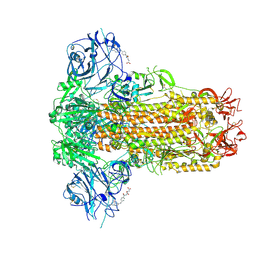 | |
9JMP
 
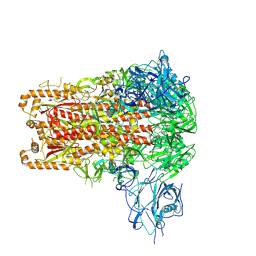 | | Cryo-EM structure of GD-BatCoV (BtCoV/Ii/GD/2014-422) S-trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yuan, H, Xiong, X, Gao, X, Li, Z, Wang, J. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
9JMH
 
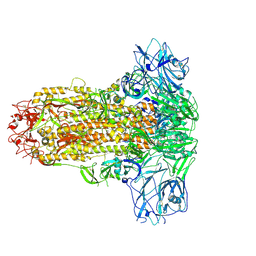 | | Cryo-EM structure of HKU25-BatCoV S-trimer stabilized with 2P and x1 disulfide bond | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Yuan, H, Xiong, X. | | Deposit date: | 2024-09-20 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and receptor binding activities of merbecovirus spike proteins reveal key signatures for human DPP4 adaptation.
Sci Adv, 11, 2025
|
|
8J4K
 
 | |
8J4L
 
 | |
7CEN
 
 | |
7CEZ
 
 | |
