3Q98
 
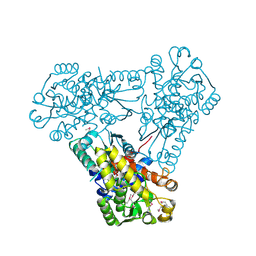 | | Structure of ygeW encoded protein from E. coli | | Descriptor: | transcarbamylase | | Authors: | Li, Y, Jing, Z, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-04 | | Last modified: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The ygeW encoded protein from Escherichia coli is a knotted ancestral catabolic transcarbamylase.
Proteins, 79, 2011
|
|
3S6K
 
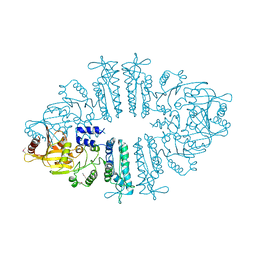 | | Crystal structure of xcNAGS | | Descriptor: | Acetylglutamate kinase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Method: | X-RAY DIFFRACTION (2.8018 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6G
 
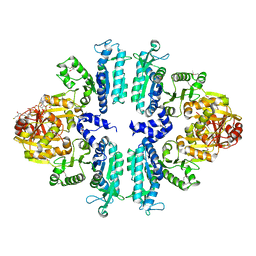 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6681 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6H
 
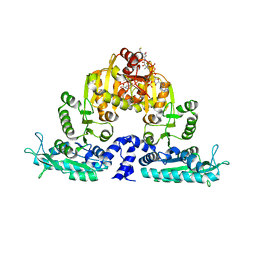 | | Crystal structure of native mmNAGS/k | | Descriptor: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S7Y
 
 | | Crystal structure of mmNAGS in Space Group P3121 at 4.3 A resolution | | Descriptor: | N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-27 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4.3077 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3TXX
 
 | | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis | | Descriptor: | Putrescine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Zhao, G, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-09-23 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of putrescine transcarbamylase from Enterococcus faecalis: Structural insights into the oligomeric assembly and the active site
To be Published
|
|
3J9X
 
 | | A Virus that Infects a Hyperthermophile Encapsidates A-Form DNA | | Descriptor: | DNA, coat protein | | Authors: | DiMaio, F, Yu, X, Rensen, E, Krupovic, M, Prangishvili, D, Egelman, E. | | Deposit date: | 2015-03-21 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A virus that infects a hyperthermophile encapsidates A-form DNA.
Science, 348, 2015
|
|
3J89
 
 | | Structural Plasticity of Helical Nanotubes Based on Coiled-Coil Assemblies | | Descriptor: | synthetic peptide | | Authors: | Egelman, E.H, Xu, C, DiMaio, F, Magnotti, E, Modlin, C, Yu, X, Wright, E, Baker, D, Conticello, V.P. | | Deposit date: | 2014-10-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural plasticity of helical nanotubes based on coiled-coil assemblies.
Structure, 23, 2015
|
|
3J9R
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3J9Q
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath, tube | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3KZC
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase | | Descriptor: | N-acetylornithine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3KZM
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZO
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with carbamyl phosphate and N-acetyl-L-norvaline | | Descriptor: | GLYCEROL, N-ACETYL-L-NORVALINE, N-acetylornithine carbamoyltransferase, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
3KZK
 
 | | Crystal structure of acetylornithine transcarbamylase complexed with acetylcitrulline | | Descriptor: | (S)-2-ACETAMIDO-5-UREIDOPENTANOIC ACID, N-acetylornithine carbamoyltransferase, SULFATE ION | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria.
J.Biol.Chem., 280, 2005
|
|
3KZN
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with N-acetyl-L-ornirthine | | Descriptor: | GLYCEROL, N-acetylornithine carbamoyltransferase, N~2~-ACETYL-L-ORNITHINE, ... | | Authors: | Shi, D, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of N-acetylornithine transcarbamoylase from Xanthomonas campestris complexed with substrates and substrate analogs imply mechanisms for substrate binding and catalysis.
Proteins, 64, 2006
|
|
5T7U
 
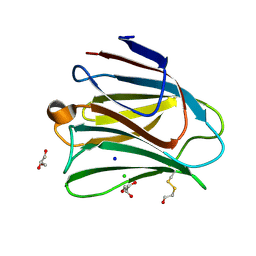 | | Crystal structure of galectin-8N in complex with Glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, Galectin-8, ... | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5T7S
 
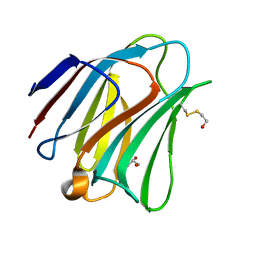 | | Crystal structure of galectin-8N in complex with Lactose | | Descriptor: | GLYCEROL, Galectin-8, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5T7I
 
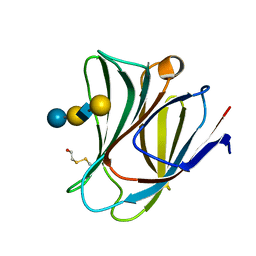 | | Crystal structure of galectin-8N in complex with LNnT | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5T7T
 
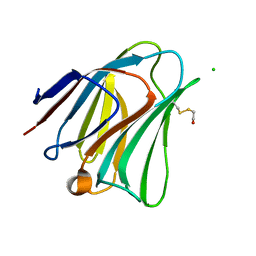 | | Galectin-8 N terminal domain in complex with LNT | | Descriptor: | CHLORIDE ION, Galectin-8, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bohari, M.H, Yu, X, Blanchard, H. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-based rationale for differential recognition of lacto- and neolacto- series glycosphingolipids by the N-terminal domain of human galectin-8.
Sci Rep, 6, 2016
|
|
5U0S
 
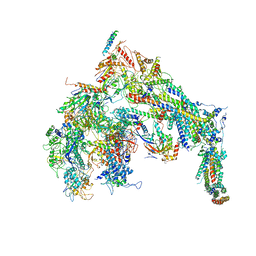 | | Cryo-EM structure of the Mediator-RNAPII complex | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
3M4J
 
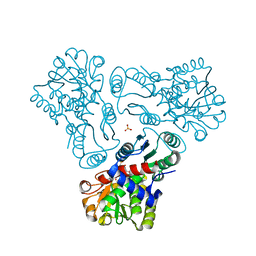 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible Post-Translational Carboxylation Modulates the Enzymatic Activity of N-Acetyl-l-ornithine Transcarbamylase.
Biochemistry, 49, 2010
|
|
3M4N
 
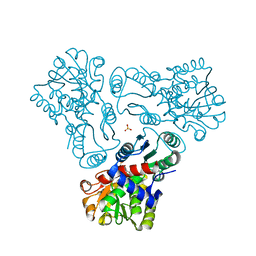 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302A mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-11 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
5U0P
 
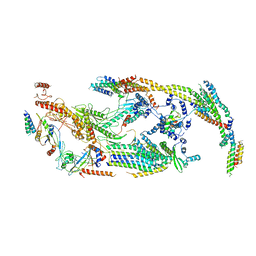 | | Cryo-EM structure of the transcriptional Mediator | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
3M5D
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302R mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
3M5C
 
 | | Crystal structure of N-acetyl-L-ornithine transcarbamylase K302E mutant complexed with PALAO | | Descriptor: | N-acetylornithine carbamoyltransferase, N~2~-acetyl-N~5~-(phosphonoacetyl)-L-ornithine, SULFATE ION | | Authors: | Li, Y, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2010-03-12 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible post-translational carboxylation modulates the enzymatic activity of N-acetyl-L-ornithine transcarbamylase.
Biochemistry, 49, 2010
|
|
