6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
4ZLS
 
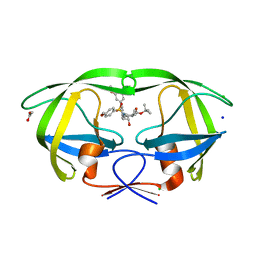 | | HIV-1 wild Type protease with GRL-096-13A (a Boc-derivative P2-Ligand, 3,-5-dimethylbiphenyl P1-Ligand) | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-05-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Potent HIV-1 Protease Inhibitors with Modified P1-Biphenyl Ligands: Synthesis, Biological Evaluation, and Enzyme-Inhibitor X-ray Structural Studies.
J.Med.Chem., 58, 2015
|
|
4RP9
 
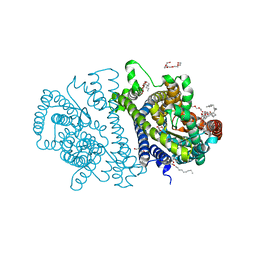 | | Bacterial vitamin C transporter UlaA/SgaT in C2 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, PENTAETHYLENE GLYCOL, ... | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2022-08-24 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RP8
 
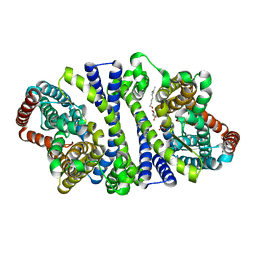 | | Bacterial vitamin C transporter UlaA/SgaT in P21 form | | Descriptor: | ASCORBIC ACID, Ascorbate-specific permease IIC component UlaA, nonyl beta-D-glucopyranoside | | Authors: | Wang, J.W. | | Deposit date: | 2014-10-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | Crystal structure of a phosphorylation-coupled vitamin C transporter.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6KA2
 
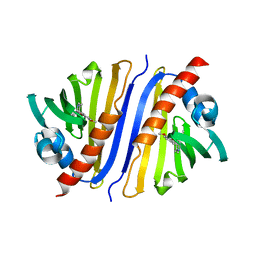 | | Crystal structure of a Thebaine synthase from Papaver somniferum in complex with TBN | | Descriptor: | (4R,7aR,12bS)-7,9-dimethoxy-3-methyl-2,4,7a,13-tetrahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
3G0E
 
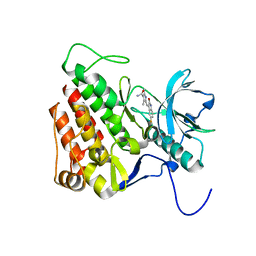 | | KIT kinase domain in complex with sunitinib | | Descriptor: | Mast/stem cell growth factor receptor, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Gajiwala, K.S, Wu, J.C, Lunney, E.A, Gemetri, G.D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FRZ
 
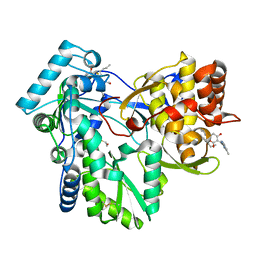 | | Crystal Structure of HCV NS5B RNA polymerase in complex with PF868554 | | Descriptor: | (6R)-6-cyclopentyl-6-[2-(2,6-diethylpyridin-4-yl)ethyl]-3-[(5,7-dimethyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl]-4-hydroxy-5,6-dihydro-2H-pyran-2-one, BETA-MERCAPTOETHANOL, N-[(benzyloxy)carbonyl]-L-alpha-glutamyl-N-[(1S)-4-oxo-4-phenyl-1-propylbut-2-en-1-yl]-L-phenylalaninamide, ... | | Authors: | Parge, H.E. | | Deposit date: | 2009-01-08 | | Release date: | 2009-03-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4-hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor.
J.Med.Chem., 52, 2009
|
|
3G0F
 
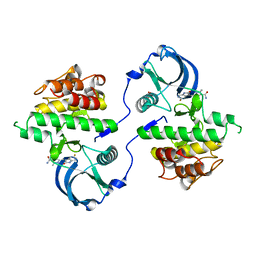 | | KIT kinase domain mutant D816H in complex with sunitinib | | Descriptor: | Mast/stem cell growth factor receptor, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide, SULFATE ION | | Authors: | Gajiwala, K.S, Wu, J.C, Lunney, E.A, Demetri, G.D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6KRG
 
 | | Crystal structure of sfGFP Y182TMSiPhe | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2019-08-21 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
3UDW
 
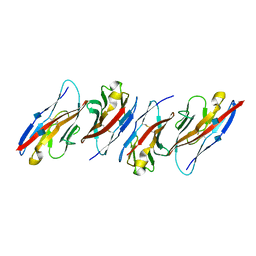 | | Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Rouge, L, Stengel, K.F, Yin, J.P, Bazan, F.J, Wiesmann, C. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UCR
 
 | | Crystal structure of the immunoreceptor TIGIT IgV domain | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Yin, J.P, Stengel, K.F, Rouge, L, Bazan, J.F, Wiesmann, C. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-14 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.627 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6MS7
 
 | | Peroxisome proliferator-activated receptor gamma ligand binding domain in complex with a novel selective PPAR-gamma modulator VSP-77 | | Descriptor: | PGC1 LXXLL motif, Peroxisome proliferator-activated receptor gamma, {[(1S)-1-(4-chlorophenyl)octyl]oxy}acetic acid | | Authors: | Yi, W, Jiang, H, Zhou, X.E, Shi, J, Zhao, G, Zhang, X, Sun, Y, Suino-Powell, K, Li, J, Li, J, Melcher, K, Xu, H.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Identification and structural insight of an effective PPAR gamma modulator with improved therapeutic index for anti-diabetic drug discovery.
Chem Sci, 11, 2020
|
|
7U2E
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADI-55688 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-55688 heavy chain, ADI-55688 light chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7U2D
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody ADG20 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADG20 heavy chain, ADG20 light chain, ... | | Authors: | Zhu, X, Yuan, M, Wilson, I.A. | | Deposit date: | 2022-02-23 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | A broad and potent neutralization epitope in SARS-related coronaviruses.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2MB9
 
 | | Human Bcl10 CARD | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | Zheng, C, Bracken, C, Wu, H. | | Deposit date: | 2013-07-26 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Architecture of the CARMA1/Bcl10/MALT1 Signalosome: Nucleation-Induced Filamentous Assembly.
Mol.Cell, 51, 2013
|
|
2P3I
 
 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P3K
 
 | | Crystal structure of Rhesus rotavirus VP8* at 100K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
2P39
 
 | | Crystal structure of human FGF23 | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor 23 | | Authors: | Mohammadi, M. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members.
Mol.Cell.Biol., 27, 2007
|
|
2P3J
 
 | |
2P23
 
 | | Crystal structure of human FGF19 | | Descriptor: | Fibroblast growth factor 19 | | Authors: | Mohammadi, M. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members.
Mol.Cell.Biol., 27, 2007
|
|
2PIE
 
 | |
6KA3
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum | | Descriptor: | PALMITIC ACID, SULFATE ION, Thebaine synthase 2 | | Authors: | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-06-20 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
5FHY
 
 | | Crystal structure of FliD (HAP2) from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellar hook-associated protein 2, SODIUM ION | | Authors: | Postel, S, Bonsor, D, Diederichs, K, Sundberg, E.J. | | Deposit date: | 2015-12-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Bacterial flagellar capping proteins adopt diverse oligomeric states.
Elife, 5, 2016
|
|
5GWV
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with a substrate analogue | | Descriptor: | (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
