6W6I
 
 | |
6P6G
 
 | | Co-crystal Structure of human SMYD3 with Isoxazole Amides Inhibitors | | Descriptor: | 5-cyclopropyl-N-{1-[({trans-4-[(4,4,4-trifluorobutyl)amino]cyclohexyl}methyl)sulfonyl]piperidin-4-yl}-1,2-oxazole-3-carboxamide, GLYCEROL, Histone-lysine N-methyltransferase SMYD3, ... | | Authors: | Elkins, P.A, Wang, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Isoxazole Amides as Potent and Selective SMYD3 Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6P6K
 
 | |
6P7Z
 
 | |
6PAF
 
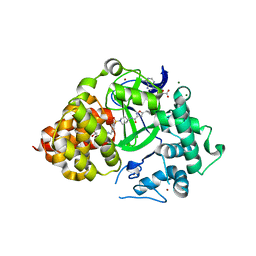 | |
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-27 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6PYH
 
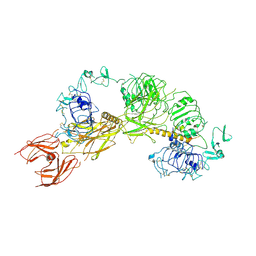 | | Cryo-EM structure of full-length IGF1R-IGF1 complex. Only the extracellular region of the complex is resolved. | | Descriptor: | Insulin-like growth factor 1 receptor, Insulin-like growth factor I | | Authors: | Li, J, Choi, E, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the activation of type 1 insulin-like growth factor receptor.
Nat Commun, 10, 2019
|
|
6PXW
 
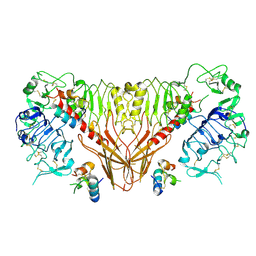 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the top part of the receptor complex. | | Descriptor: | Insulin, Insulin receptor | | Authors: | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | Deposit date: | 2019-07-28 | | Release date: | 2019-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
6VLB
 
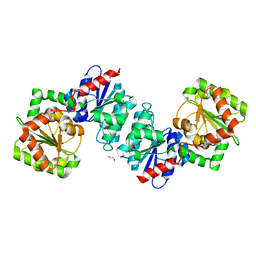 | |
6VLC
 
 | |
6WML
 
 | | Human TLR8 bound to the potent agonist, GS-9688 (Selgantolimod) | | Descriptor: | (2R)-2-[(2-amino-7-fluoropyrido[3,2-d]pyrimidin-4-yl)amino]-2-methylhexan-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 8, ... | | Authors: | Appleby, T.C, Perry, J.K, Mish, M, Villasenor, A.G, Mackman, R.L. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of GS-9688 (Selgantolimod) as a Potent and Selective Oral Toll-Like Receptor 8 Agonist for the Treatment of Chronic Hepatitis B.
J.Med.Chem., 63, 2020
|
|
6XTG
 
 | | Ab 1116NS19.9 bound to CA19-9 | | Descriptor: | Heavy chain, Light chain, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUN
 
 | | Ab 5b1 bound to CA19-9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUK
 
 | | AbLIFT design 15 of Ab 1116NS19.9 | | Descriptor: | GLYCEROL, Heavy chain, Light chain, ... | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUD
 
 | | Apo Ab 1116NS19.9 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
6XUL
 
 | | Apo Ab 5b1 | | Descriptor: | Heavy chain, Light chain | | Authors: | Diskin, R, Borenstein-Katz, A. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Biomolecular Recognition of the Glycan Neoantigen CA19-9 by Distinct Antibodies.
J.Mol.Biol., 433, 2021
|
|
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
1NLP
 
 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL2 (MN8-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
1NLO
 
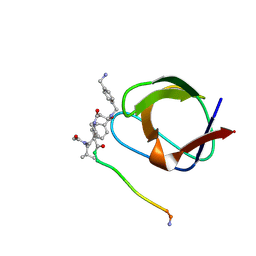 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | C-SRC, NL1 (MN7-MN2-MN1-PLPPLP) | | Authors: | Feng, S, Kapoor, T.M, Shirai, F, Combs, A.P, Schreiber, S.L. | | Deposit date: | 1996-08-04 | | Release date: | 1997-01-27 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the binding of SH3 ligands with non-peptide elements identified by combinatorial synthesis.
Chem.Biol., 3, 1996
|
|
8IRI
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 5-millisecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRD
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 20-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IR6
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 20-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRG
 
 | | XFEL structure of cyanobacterial photosystem II following two flashes (2F) with a 30-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IR7
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-nanosecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
8IRA
 
 | | XFEL structure of cyanobacterial photosystem II following one flash (1F) with a 200-microsecond delay | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Suga, M, Shen, J.R. | | Deposit date: | 2023-03-17 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Oxygen-evolving photosystem II structures during S 1 -S 2 -S 3 transitions.
Nature, 626, 2024
|
|
