1VGO
 
 | | Crystal Structure of Archaerhodopsin-2 | | 分子名称: | Archaerhodopsin 2, RETINAL, SULFATE ION, ... | | 著者 | Yoshimura, K, Enami, N, Murakami, M, Okumura, H, Ihara, K, Kouyama, T. | | 登録日 | 2004-04-28 | | 公開日 | 2005-10-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of archaerhodopsin-1 and -2: Common structural motif in archaeal light-driven proton pumps
J.Mol.Biol., 358, 2006
|
|
2Z55
 
 | |
7DHW
 
 | | Crystal structure of myosin-XI motor domain in complex with ADP-ALF4 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Suzuki, K, Haraguchi, T, Tamanaha, M, Yoshimura, K, Imi, T, Tominaga, M, Sakayama, H, Nishiyama, T, Ito, K, Murata, T. | | 登録日 | 2020-11-17 | | 公開日 | 2021-05-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of ultrafast myosin, its amino acid sequence, and structural features.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2EI4
 
 | | Trimeric complex of archaerhodopsin-2 | | 分子名称: | 2,3-DI-PHYTANYL-GLYCEROL, Archaerhodopsin-2, BACTERIORUBERIN, ... | | 著者 | Kouyama, T. | | 登録日 | 2007-03-11 | | 公開日 | 2008-01-01 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural role of bacterioruberin in the trimeric structure of archaerhodopsin-2
J.Mol.Biol., 375, 2008
|
|
9II9
 
 | |
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | 分子名称: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-14 | | 公開日 | 2022-09-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8GS6
 
 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | 登録日 | 2022-09-05 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.86 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
8IF2
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Kimura, K, Suzuki, T, Hashiguchi, T. | | 登録日 | 2023-02-17 | | 公開日 | 2023-05-17 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
8IOS
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOU
 
 | | Structure of SARS-CoV-2 XBB.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOV
 
 | | Structure of SARS-CoV-2 XBB.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
8IOT
 
 | | Structure of the SARS-CoV-2 XBB.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Anraku, Y, Kita, S, Yajima, H, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | 登録日 | 2023-03-13 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants.
Nat Commun, 14, 2023
|
|
3NTC
 
 | | Crystal structure of KD-247 Fab, an anti-V3 antibody that inhibits HIV-1 Entry | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Fab heavy chain, ... | | 著者 | Sarafianos, S.G, Kirby, K.A. | | 登録日 | 2010-07-03 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural basis of clade-specific HIV-1 neutralization by humanized anti-V3 monoclonal antibody KD-247.
Faseb J., 29, 2015
|
|
8WMD
 
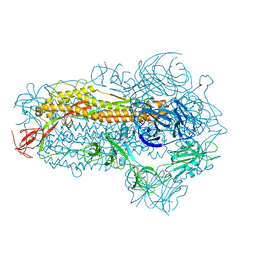 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.71 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8WMF
 
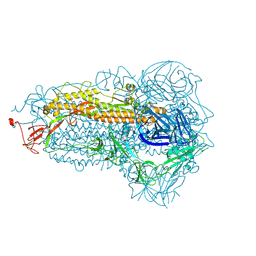 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-10-03 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.51 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XLM
 
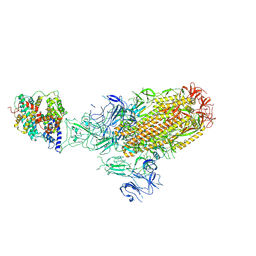 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-11-27 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8XLN
 
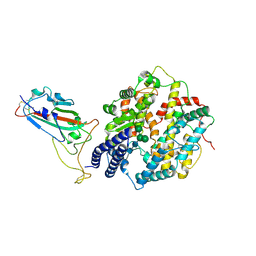 | | Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | 登録日 | 2023-12-26 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant.
Microbiol Immunol, 68, 2024
|
|
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | 分子名称: | ORF1a polyprotein | | 著者 | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2023-10-07 | | 公開日 | 2024-01-24 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2023-10-06 | | 公開日 | 2023-12-13 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | 著者 | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2023-10-07 | | 公開日 | 2023-12-20 | | 最終更新日 | 2025-02-19 | | 実験手法 | X-RAY DIFFRACTION (2.067 Å) | | 主引用文献 | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Suzuki, T, Kimura, K, Hashiguchi, T. | | 登録日 | 2022-05-26 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.36 Å) | | 主引用文献 | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
1IYN
 
 | |
2ZFE
 
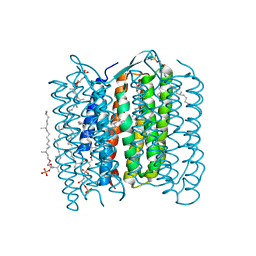 | | Crystal structure of bacteriorhodopsin-xenon complex | | 分子名称: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, ... | | 著者 | Kouyama, T. | | 登録日 | 2007-12-31 | | 公開日 | 2008-11-11 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Effect of xenon binding to a hydrophobic cavity on the proton pumping cycle in bacteriorhodopsin
J.Mol.Biol., 384, 2008
|
|
