7CE1
 
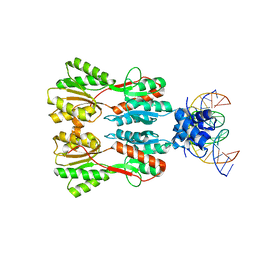 | | Complex STRUCTURE OF TRANSCRIPTION FACTOR SghR with its COGNATE DNA | | 分子名称: | LacI-type transcription factor, promoter DNA | | 著者 | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | 登録日 | 2020-06-21 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDV
 
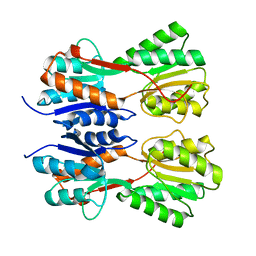 | | STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR | | 分子名称: | LacI-type transcription factor | | 著者 | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
7CDX
 
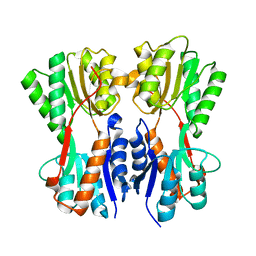 | | Complex STRUCTURE OF A NOVEL VIRULENCE REGULATION FACTOR SghR with its effector sucrose | | 分子名称: | LacI-type transcription factor, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Ye, F.Z, Wang, C, Yan, X.F, Zhang, L.H, Gao, Y.G. | | 登録日 | 2020-06-20 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Structural basis of a novel repressor, SghR, controllingAgrobacteriuminfection by cross-talking to plants.
J.Biol.Chem., 295, 2020
|
|
4EMM
 
 | |
8X84
 
 | |
8X83
 
 | |
8X82
 
 | |
6RJK
 
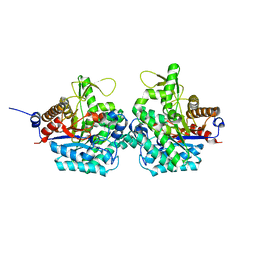 | | Structure of virulence factor SghA from Agrobacterium tumefaciens | | 分子名称: | Beta-glucosidase | | 著者 | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | 登録日 | 2019-04-27 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RK2
 
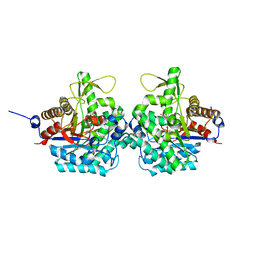 | | Complex structure of virulence factor SghA mutant with its substrate SAG | | 分子名称: | 2-(alpha-L-altropyranosyloxy)benzoic acid, Beta-glucosidase | | 著者 | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | 登録日 | 2019-04-30 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJM
 
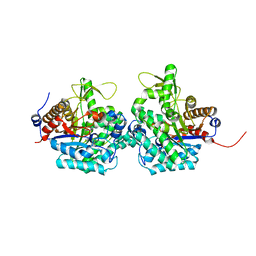 | | Complex structure of virulence factor SghA and its hydrolysis product glucose | | 分子名称: | Beta-glucosidase, alpha-D-glucopyranose | | 著者 | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | 登録日 | 2019-04-27 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6RJO
 
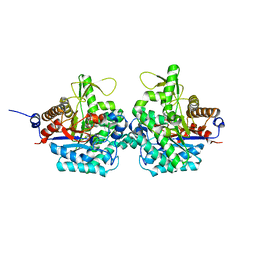 | | Complex structure of virulence factor SghA with its substrate analog salicin | | 分子名称: | 2-(hydroxymethyl)phenyl beta-D-glucopyranoside, Beta-glucosidase | | 著者 | Ye, F.Z, Wang, C, Chang, C.Q, Zhang, L.H, Gao, Y.G. | | 登録日 | 2019-04-28 | | 公開日 | 2019-10-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.804 Å) | | 主引用文献 | Agrobacteria reprogram virulence gene expression by controlled release of host-conjugated signals.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7WKS
 
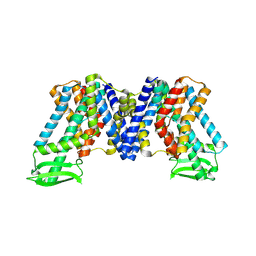 | | Apo state of AtPIN3 | | 分子名称: | Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-01-11 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
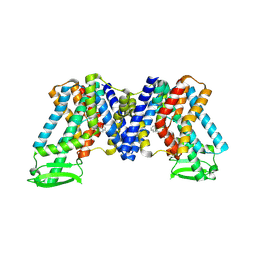 | | NPA bound state of AtPIN3 | | 分子名称: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-01-11 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7XXB
 
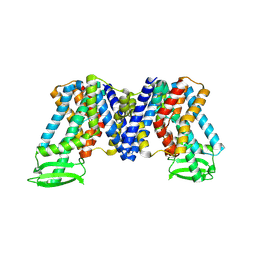 | | IAA bound state of AtPIN3 | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, Auxin efflux carrier component 3 | | 著者 | Su, N. | | 登録日 | 2022-05-29 | | 公開日 | 2022-08-10 | | 最終更新日 | 2022-09-28 | | 実験手法 | ELECTRON MICROSCOPY (2.93 Å) | | 主引用文献 | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
6L1Q
 
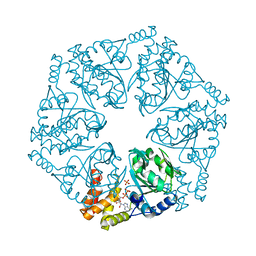 | | Crystal structure of AfCbbQ2, a MoxR AAA+-ATPase and CbbQO-type Rubisco activase from Acidithiobacillus ferrooxidans | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CbbQ protein, PHOSPHATE ION | | 著者 | Ye, F.Z, Tsai, Y.C.C, Mueller-Cajar, O, Gao, Y.G. | | 登録日 | 2019-09-30 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Insights into the mechanism and regulation of the CbbQO-type Rubisco activase, a MoxR AAA+ ATPase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8JMA
 
 | |
8JMH
 
 | |
8JMI
 
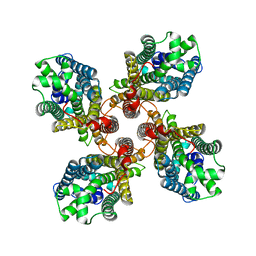 | |
8JM9
 
 | |
8JME
 
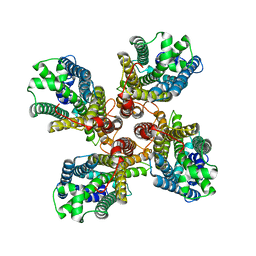 | |
3F7O
 
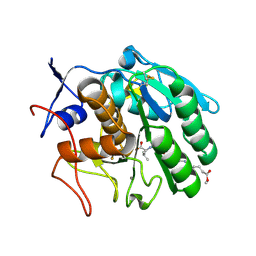 | | Crystal structure of Cuticle-Degrading Protease from Paecilomyces lilacinus (PL646) | | 分子名称: | (MSU)(ALA)(ALA)(PRO)(VAL), CALCIUM ION, Serine protease | | 著者 | Liang, L, Lou, Z, Meng, Z, Rao, Z, Zhang, K. | | 登録日 | 2008-11-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The crystal structures of two cuticle-degrading proteases from nematophagous fungi and their contribution to infection against nematodes.
Faseb J., 24, 2010
|
|
6GH5
 
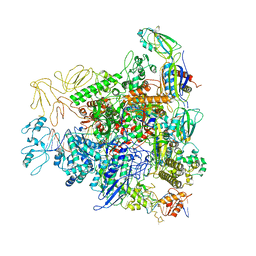 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme transcription open complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-04 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GFW
 
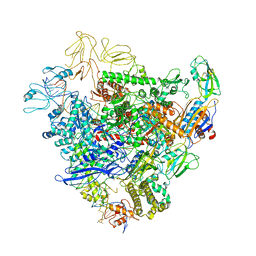 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme initial transcribing complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-02 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
6GH6
 
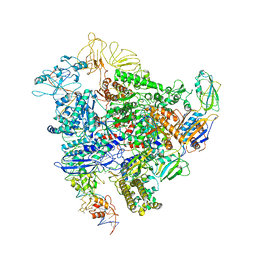 | | Cryo-EM structure of bacterial RNA polymerase-sigma54 holoenzyme intermediate partially loaded complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Glyde, R, Ye, F.Z, Zhang, X.D. | | 登録日 | 2018-05-04 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Structures of Bacterial RNA Polymerase Complexes Reveal the Mechanism of DNA Loading and Transcription Initiation.
Mol. Cell, 70, 2018
|
|
8REB
 
 | |
