5ZRU
 
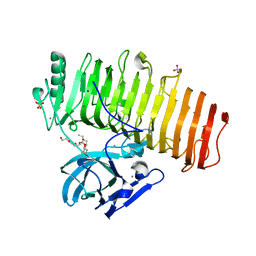 | | Crystal structure of Agl-KA catalytic domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Yano, S, Makabe, K. | | Deposit date: | 2018-04-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal structure of the catalytic unit of GH 87-type alpha-1,3-glucanase Agl-KA from Bacillus circulans.
Sci Rep, 9, 2019
|
|
6K0U
 
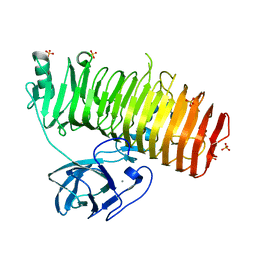 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0M
 
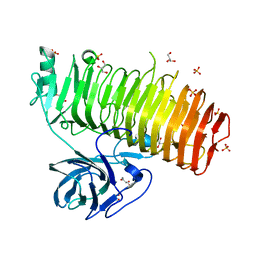 | | Catalytic domain of GH87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11 | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, GLYCEROL, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0Q
 
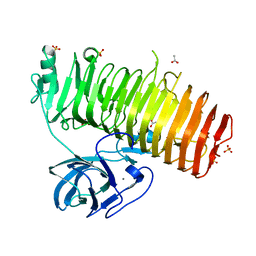 | | Catalytic domain of GH87 alpha-1,3-glucanase D1068A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0N
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0P
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1045A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.424 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0V
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with tetrasaccharides | | Descriptor: | Alpha-1,3-glucanase, CALCIUM ION, SULFATE ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
6K0S
 
 | | Catalytic domain of GH87 alpha-1,3-glucanase D1069A in complex with nigerose | | Descriptor: | ACETIC ACID, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Itoh, T, Intuy, R, Suyotha, W, Hayashi, J, Yano, S, Makabe, K, Wakayama, M, Hibi, T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Structural insights into substrate recognition and catalysis by glycoside hydrolase family 87 alpha-1,3-glucanase from Paenibacillus glycanilyticus FH11.
Febs J., 287, 2020
|
|
7C7D
 
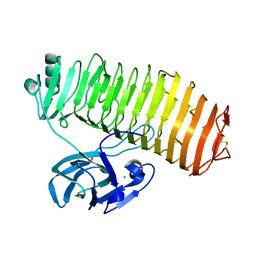 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
1IRA
 
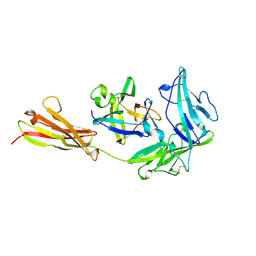 | | COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-1 RECEPTOR, INTERLEUKIN-1 RECEPTOR ANTAGONIST | | Authors: | Schreuder, H.A, Tardif, C, Tramp-Kalmeyer, S, Soffientini, A, Sarubbi, E, Akeson, A, Bowlin, T, Yanofsky, S, Barrett, R.W. | | Deposit date: | 1998-04-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Nature, 386, 1997
|
|
5AXI
 
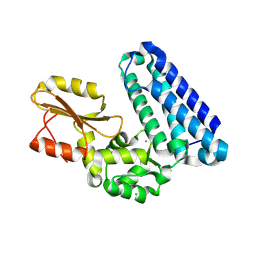 | | Crystal structure of Cbl-b TKB domain in complex with Cblin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cblin, ... | | Authors: | Ohno, A, Maita, N, Ochi, A, Nakao, R, Nikawa, T. | | Deposit date: | 2015-07-29 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the TKB domain of ubiquitin ligase Cbl-b complexed with its small inhibitory peptide, Cblin
Arch.Biochem.Biophys., 594, 2016
|
|
7BV9
 
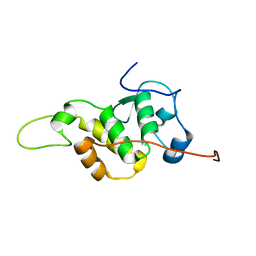 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
8YKI
 
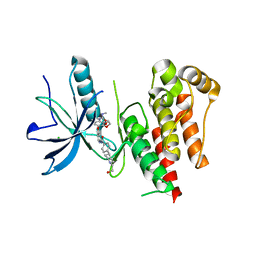 | | FGFR-1 in complex with ligand tasurgratinib | | Descriptor: | CHLORIDE ION, Fibroblast growth factor receptor 1, Tasurgratinib | | Authors: | Ikemori-Kawada, M, Watanabe Miyano, S. | | Deposit date: | 2024-03-05 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Antitumor Activity of Tasurgratinib as an Orally Available FGFR1-3 Inhibitor in Cholangiocarcinoma Models With FGFR2-fusion.
Anticancer Res., 44, 2024
|
|
6CG7
 
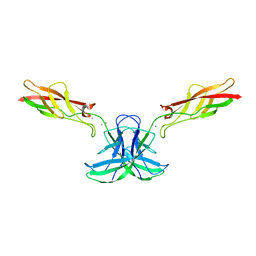 | | mouse cadherin-22 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-22 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGS
 
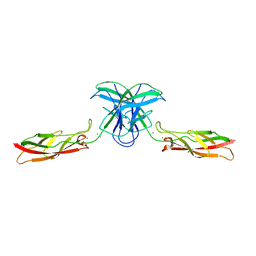 | | mouse cadherin-7 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-7, GLYCEROL | | Authors: | Brasch, J, Harrison, O.J, Kaczynska, A, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
1XMZ
 
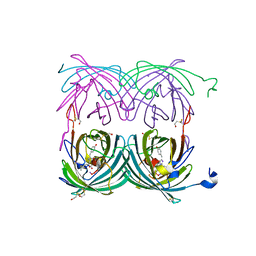 | | Crystal structure of the dark state of kindling fluorescent protein kfp from anemonia sulcata | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Quillin, M.L, Anstrom, D.M, Shu, X, O'Leary, S, Kallio, K, Chudakov, D.M, Remington, S.J. | | Deposit date: | 2004-10-04 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Kindling Fluorescent Protein from Anemonia sulcata: Dark-State Structure at 1.38 Resolution
Biochemistry, 44, 2005
|
|
6YPE
 
 | |
6CG6
 
 | | mouse cadherin-10 EC1-2 adhesive fragment | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cadherin-10, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGU
 
 | | mouse cadherin-6 EC1-2 adhesive fragment | | Descriptor: | CALCIUM ION, Cadherin-6 | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2018-02-20 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
6CGB
 
 | | chimera of mouse cadherin-11 EC1 and mouse cadherin-6 EC2 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-11, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L, Kaeser, B. | | Deposit date: | 2018-02-19 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Homophilic and Heterophilic Interactions of Type II Cadherins Identify Specificity Groups Underlying Cell-Adhesive Behavior.
Cell Rep, 23, 2018
|
|
4HE4
 
 | |
4EDS
 
 | |
4EDO
 
 | |
2KUS
 
 | |
1ILR
 
 | |
