6P8Y
 
 | |
6P8X
 
 | |
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | 分子名称: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | 著者 | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | 登録日 | 2006-07-14 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | 分子名称: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | 著者 | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | 登録日 | 2022-10-08 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5F
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) L167F Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
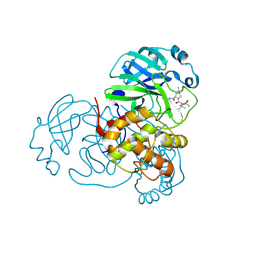 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
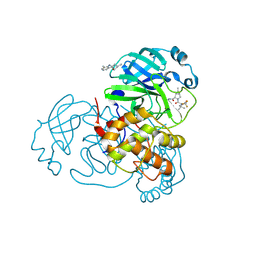 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7K
 
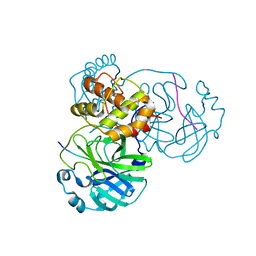 | |
8H5P
 
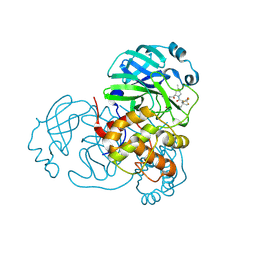 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6I
 
 | | The crystal structure of SARS-CoV-2 3C-like protease Double Mutant (L50F and E166V) in complex with a traditional Chinese Medicine Inhibitors | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-12 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HBK
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M. | | 登録日 | 2022-10-29 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
6DUM
 
 | | ALDH1A1 N121S in complex with 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one (compound 13g) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-2-(oxetan-3-yl)-5-phenyl-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | 著者 | Buchman, C.D, Hurley, T.D. | | 登録日 | 2018-06-21 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-Based Optimization of a Novel Class of Aldehyde Dehydrogenase 1A (ALDH1A) Subfamily-Selective Inhibitors as Potential Adjuncts to Ovarian Cancer Chemotherapy.
J.Med.Chem., 61, 2018
|
|
7L05
 
 | | Complex of novel maytansinoid M24 bound to T2R-TTL (two tubulin alpha/beta heterodimers, RB3 stathmin-like domain, and tubulin tyrosine ligase) | | 分子名称: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methylamino)propanoyl]amino}propanoate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Franklin, M.C. | | 登録日 | 2020-12-11 | | 公開日 | 2021-12-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | A Biparatopic Antibody-Drug Conjugate to Treat MET-Expressing Cancers, Including Those that Are Unresponsive to MET Pathway Blockade.
Mol.Cancer Ther., 20, 2021
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | 分子名称: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | 著者 | Hong, W, Chen, H, Wu, Q, Lin, T. | | 登録日 | 2019-09-23 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (4.45 Å) | | 主引用文献 | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8W1N
 
 | |
3S2A
 
 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | 分子名称: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | 著者 | Whittington, D.A, Tang, J, Yakowec, P. | | 登録日 | 2011-05-16 | | 公開日 | 2011-06-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
6LN1
 
 | | A natural inhibitor of DYRK1A for treatment of diabetes mellitus | | 分子名称: | 1,3,5,8-tetrakis(oxidanyl)xanthen-9-one, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | 著者 | Li, H, Chen, L.X, Zheng, M.Z, Zhang, Q.Z, Zhang, C.L, Wu, C.R, Yang, K.Y, Song, Z.R, Wang, Q.Q, Li, C, Zhou, Y.R, Chen, J.C. | | 登録日 | 2019-12-28 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.699 Å) | | 主引用文献 | A natural DYRK1A inhibitor as a potential stimulator for beta-cell proliferation in diabetes.
Clin Transl Med, 11, 2021
|
|
7D7L
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in complex with YM155 | | 分子名称: | 1-(2-methoxyethyl)-2-methyl-3-(pyrazin-2-ylmethyl)benzo[f]benzimidazol-3-ium-4,9-dione, CAFFEINE, GLYCEROL, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | 分子名称: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | 著者 | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | 登録日 | 2020-10-04 | | 公開日 | 2021-04-21 | | 最終更新日 | 2021-11-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
