1CZ9
 
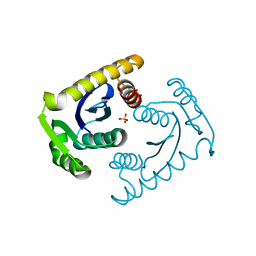 | | ATOMIC RESOLUTION ASV INTEGRASE CORE DOMAIN (D64N) FROM CITRATE | | Descriptor: | AVIAN SARCOMA VIRUS INTEGRASE, CITRIC ACID, SULFATE ION | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Merkel, G, Skalka, A.M, Wlodawer, A. | | Deposit date: | 1999-09-01 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
1CXU
 
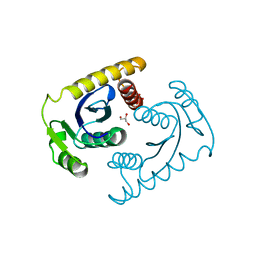 | | 1.42A RESOLUTION ASV INTEGRASE CORE DOMAIN FROM CITRATE | | Descriptor: | CITRIC ACID, GLYCEROL, PROTEIN (AVIAN SARCOMA VIRUS INTEGRASE) | | Authors: | Lubkowski, J, Dauter, Z, Yang, F, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1999-08-30 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Atomic resolution structures of the core domain of avian sarcoma virus integrase and its D64N mutant.
Biochemistry, 38, 1999
|
|
3H94
 
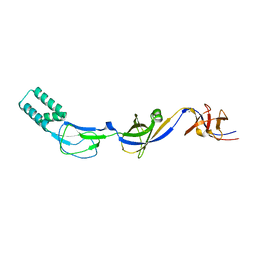 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
3HGG
 
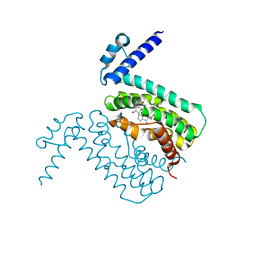 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3OOC
 
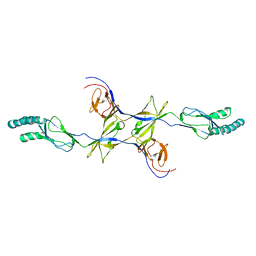 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2010-08-30 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli.
J.Mol.Biol., 393, 2009
|
|
3BCG
 
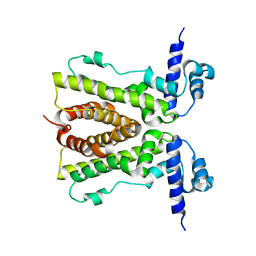 | | Conformational changes of the AcrR regulator reveal a mechanism of induction | | Descriptor: | HTH-type transcriptional regulator acrR | | Authors: | Gu, R, Li, M, Su, C.C, Long, F, Yang, F, McDermott, G, Yu, E.Y. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conformational change of the AcrR regulator reveals a possible mechanism of induction.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4NND
 
 | |
6LAE
 
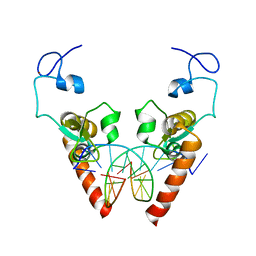 | | Crystal structure of the DNA-binding domain of human XPA in complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*TP*CP*GP*CP*CP*T)-3'), DNA (5'-D(P*TP*GP*GP*CP*GP*AP*GP*AP*TP*GP*C)-3'), DNA repair protein complementing XP-A cells, ... | | Authors: | Lian, F.M, Yang, X, Jiang, Y.L, Yang, F, Li, C, Yang, W, Qian, C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA.
Int.J.Biol.Macromol., 148, 2020
|
|
3B9L
 
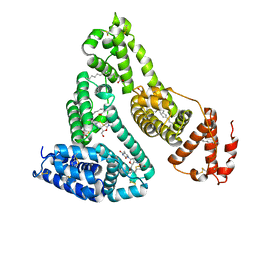 | | Human serum albumin complexed with myristate and AZT | | Descriptor: | 3'-azido-3'-deoxythymidine, MYRISTIC ACID, Serum albumin | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
3B9M
 
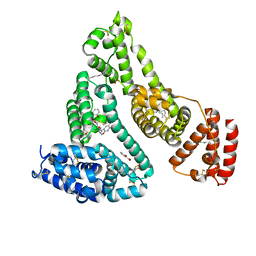 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
4GFU
 
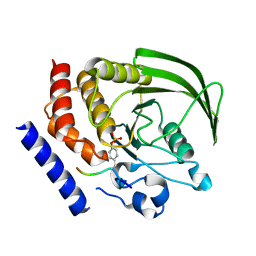 | | PTPN18 in complex with HER2-pY1248 phosphor-peptides | | Descriptor: | HER2-pY1248 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
4GFV
 
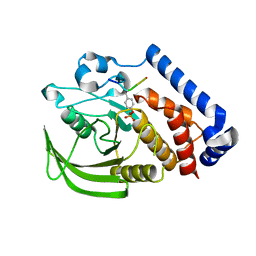 | | PTPN18 in complex with HER2-pY1196 phosphor-peptides | | Descriptor: | HER2-pY1196 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
5J8R
 
 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 12 - K61R mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Li, H, Yang, F, Xu, Y.F, Wang, W.J, Xiao, P, Yu, X, Sun, J.P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Crystal structure and substrate specificity of PTPN12.
Cell Rep, 15, 2016
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
1NDG
 
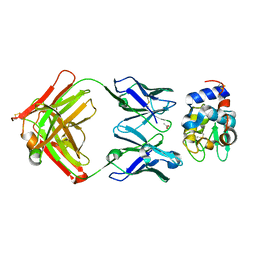 | | Crystal structure of Fab fragment of antibody HyHEL-8 complexed with its antigen lysozyme | | Descriptor: | ACETIC ACID, Lysozyme C, antibody kappa light chain, ... | | Authors: | Mariuzza, R.A, Li, Y, Li, H, Yang, F, Smith-Gill, S.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-06-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of the maturation of an antibody response to a protein antigen
Nat.Struct.Biol., 10, 2003
|
|
3HGY
 
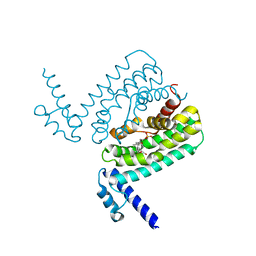 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
1R5H
 
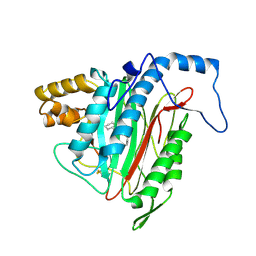 | | Crystal Structure of MetAP2 complexed with A320282 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R58
 
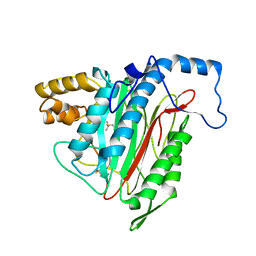 | | Crystal Structure of MetAP2 complexed with A357300 | | Descriptor: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-((2S,3R)-3-AMINO-2-HYDROXY-5-(ISOPROPYLSULFANYL)PENTANOYL)-N-3-CHLOROBENZOYL HYDRAZIDE | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Ericken, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
1R5G
 
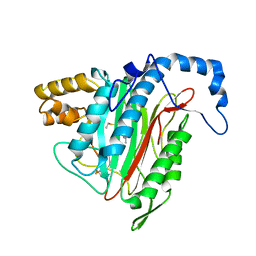 | | Crystal Structure of MetAP2 complexed with A311263 | | Descriptor: | (2S,3R)-3-AMINO-2-HYDROXY-5-(ETHYLSULFANYL)PENTANOYL-((S)-(-)-(1-NAPHTHYL)ETHYL)AMIDE, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | Deposit date: | 2003-10-10 | | Release date: | 2004-10-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
3CX9
 
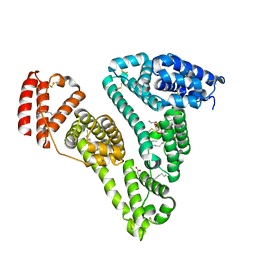 | | Crystal Structure of Human serum albumin complexed with Myristic acid and lysophosphatidylethanolamine | | Descriptor: | (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, MYRISTIC ACID, Serum albumin | | Authors: | Guo, S, Yang, F, Chen, L, Bian, C, Huang, M. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of transport of lysophospholipids by human serum albumin.
Biochem.J., 423, 2009
|
|
4AUA
 
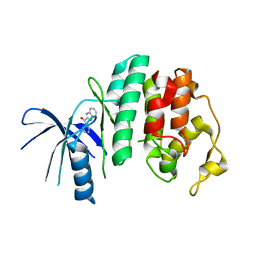 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
4Z69
 
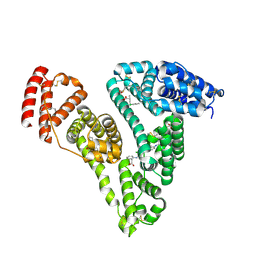 | | Human serum albumin complexed with palmitic acid and diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, PALMITIC ACID, PENTADECANOIC ACID, ... | | Authors: | Zhang, Y, Yang, F. | | Deposit date: | 2015-04-04 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis of non-steroidal anti-inflammatory drug diclofenac binding to human serum albumin.
Chem.Biol.Drug Des., 86, 2015
|
|
5YUD
 
 | | Flagellin derivative in complex with the NLR protein NAIP5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Baculoviral IAP repeat-containing protein 1e, Phase 2 flagellin,Flagellin | | Authors: | Yang, X.R, Yang, F, Wang, W.G, Lin, G.Z. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for specific flagellin recognition by the NLR protein NAIP5.
Cell Res., 28, 2018
|
|
5GOR
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 | | Descriptor: | Alkaline Invertase, GLYCEROL, SULFATE ION | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
