8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | 著者 | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | 登録日 | 2022-11-01 | | 公開日 | 2023-01-25 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (6.03 Å) | | 主引用文献 | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
7FBT
 
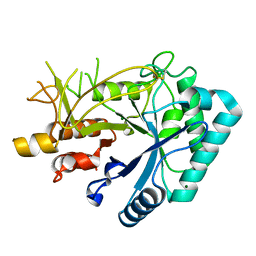 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | 分子名称: | Chitinase, MAGNESIUM ION | | 著者 | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | 登録日 | 2021-07-12 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
5ISL
 
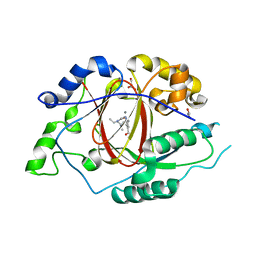 | |
5IVC
 
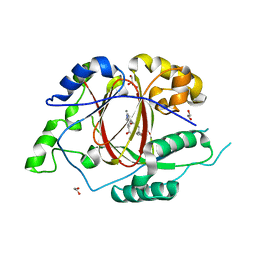 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N3 (4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid) | | 分子名称: | 1,2-ETHANEDIOL, 4'-[(2-phenylethyl)carbamoyl][2,2'-bipyridine]-4-carboxylic acid, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.573 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVB
 
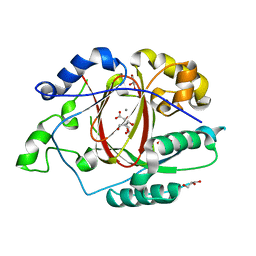 | |
5IWF
 
 | |
5IVF
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | 分子名称: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVV
 
 | |
5IVJ
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N11 [3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid] | | 分子名称: | 3-({1-[2-(4,4-difluoropiperidin-1-yl)ethyl]-5-fluoro-1H-indazol-3-yl}amino)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-21 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.569 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IVY
 
 | |
5IVE
 
 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N8 ( 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile) | | 分子名称: | 5-methyl-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.783 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5IW0
 
 | |
8H15
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.14182 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.44681 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0X
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H16
 
 | | Structure of SARS-CoV-1 Spike Protein (S/native) at pH 5.5, Open Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.35534 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H0Y
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-112 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, LINOLEIC ACID, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.85 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
6BH3
 
 | |
6BGW
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(4,4-difluoropiperidin-1-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N41) | | 分子名称: | 2-{(S)-(2-chlorophenyl)[2-(4,4-difluoropiperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.644 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 5-(1-(tert-butyl)-1H-pyrazol-4-yl)-6-isopropyl-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile (Compound N75/CPI-48) | | 分子名称: | 1,2-ETHANEDIOL, 5-(1-tert-butyl-1H-pyrazol-4-yl)-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, GLYCEROL, ... | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2017-10-29 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.047 Å) | | 主引用文献 | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
